Autumn newsletter 2022
Newsletters ·
New REST API
We are excited to announce that we have expanded our REST API so that you can now query both HGNC and VGNC data! The genenames.org REST web-service is a convenient and quick way of searching and fetching data from our database within a script/program. Users may request results as either XML or JSON making our data easier to parse. Previously this was only supported for HGNC data, but it’s now possible to search & fetch VGNC data using the same REST API. Just prefix the request type with vgnc/ and you will be searching and retrieving records within the VGNC sister site. Full details can be found at https://www.genenames.org/help/rest.
HGNC track on UCSC Genome Browser
We are delighted by the new HUGO Gene Nomenclature track that has recently been added to the UCSC Genome Browser! This new track is fully searchable and shows the HGNC approved symbol and HGNC ID along with an associated RefSeq ID, as shown in the image below.
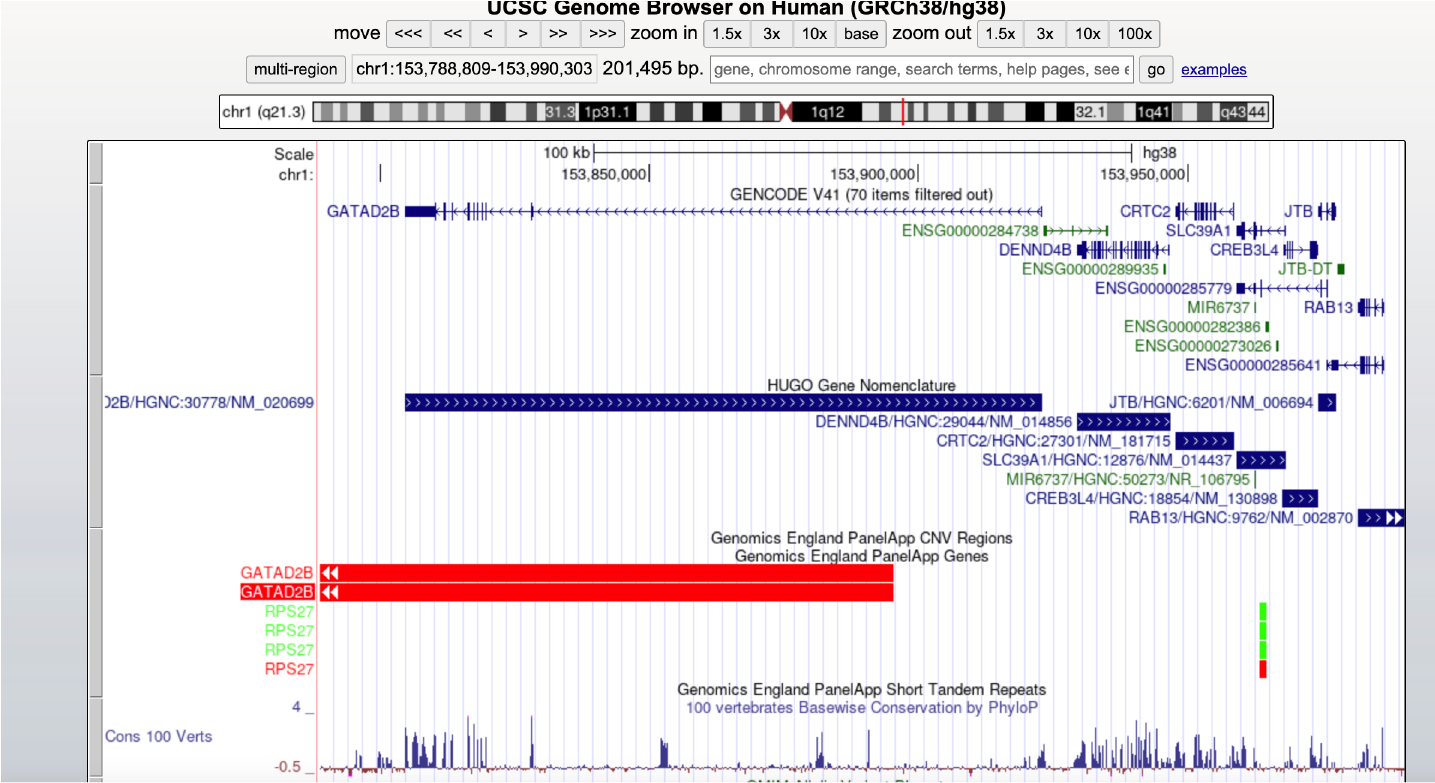
Clicking on the HGNC gene in the browser links through to a UCSC-hosted report page that includes previous and alias gene symbols and names, as well other associated information such gene IDs for associated resources like Ensembl and UniProt. Importantly, the UCSC have added a “thesaurus” to their resource which means that HGNC aliases and previous nomenclature can now be used to access the correct gene in the UCSC genome browser.
Update on genes with the ‘stable’ tag
As of 16th November 2022 we have 3027 gene symbols tagged as ‘stable’, an increase of 81 stable genes since our Summer newsletter. These include several genes named with the CD# (CD molecule) and SLC# (solute carrier) root symbols, such as: CD44, CD55, CD59, SLC1A4, SLC2A10 and SLC5A1
The stabilisation process includes a review of the gene symbol and name; in a relatively small number of cases the gene symbol may be updated before the stable tag is added. For example, HGNC:11075 was updated from SLC9A3R1 (SLC9A3 regulator 1) to NHERF1 (NHERF family PDZ scaffold protein 1) due in part to the much higher usage of the NHERF1 symbol by the community. A consultation was held with research groups to confirm support for the change. The paralogs HGNC:11076 and HGNC:19891 were likewise renamed from SLC9A3R2 and PDZD3 to NHERF2 and NHERF4, while the fourth member of the group has the approved symbol PDZK1, which is well published, so PDZK1 has been retained as the approved symbol and assigned the stable tag, while NHERF3 is a symbol alias for this gene. The new NHERF# root symbol groups together these four paralogs encoding scaffold proteins.
Updates to placeholder symbols
Within the last quarter we have been able to update the following placeholder symbols with more informative nomenclature:
- FAM183A -> CFAP144, cilia and flagella associated protein 144
- FAM102A -> EEIG1, estrogen-induced osteoclastogenesis regulator 1
- FAM102B -> EEIG2, EEIG family member 2
Gene Symbols in the News
This quarter we have some stories about how immune-related gene variants have altered our own and our ancestors’ susceptibility to disease. A new study on skeletons from the period of the black death and afterwards has found that mutations in the ERAP2 gene had a protective effect against the plague but that the same mutations are associated with auto-immune diseases such as Crohn’s disease in people today. Variants in the TYK2 gene have been shown to be protective against severe COVID-19 but to also increase the risk of auto-immune disease, particularly lupus.
In other COVID-related news, researchers found that people with a particular allele of the HLA-DQB1 gene, HLA-DQB1*06, showed an increased antibody response to vaccination against the disease than the rest of the population. The allele has a 30-40% prevalence in the UK.
Recent work into the mechanisms that result in cachexia in cancer patients has shown that the muscles of such patients have increased levels of the UBR2-encoded protein and an associated loss of myosin heavy chain. Studies in mice have shown that existing drugs can be used to reduce the UBR2 increase, raising hopes that such drugs may be used as part of cancer care in the future.
Finally, a study has suggested that the ARHGAP11B gene, a human-specific duplication of the ARHGAP11A gene, has a key role in human neocortex development. Expression of ARHGAP11B in chimpanzee cerebral organoids resulted in an increase of neocortex progenitor cells.
Meeting News
Ruth attended Genome Informatics 2022 from September 21st-23rd at the Genome Campus in Hinxton where she presented a poster based on the naming of histone genes across mammalian species. Delegates liked the images of the animals whose genomes were included in the study!
Tamsin will be attending PAG30 (30th Plant & Animal Genome Conference) in January 2023 in sunny San Diego. She will be presenting a poster entitled “Naming Vertebrate Orthologs of Human Unitary Pseudogenes” and taking part in the EMBL-EBI workshop.
Publications
Bruford EA, Braschi B, Haim-Vilmovsky L, Jones TEM, Seal RL, Tweedie S. The importance of being the HGNC. Hum Genomics. 2022 Nov 15;16(1):58. DOI: 10.1186/s40246-022-00432-w. PMID: 36380364
Dornburg A, Mallik R, Wang Z, Bernal MA, Thompson B, Bruford EA, Nebert DW, Vasiliou V, Yohe LR, Yoder JA, Townsend JP. Placing human gene families into their evolutionary context. Hum Genomics. 2022 Nov 11;16(1):56. DOI: 10.1186/s40246-022-00429-5 PMID: 36369063; PMCID: PMC965288
Seal RL, Braschi B, Gray K, Jones TEM, Tweedie S, Haim-Vilmovsky L, Bruford EA. Genenames.org: the HGNC resources in 2023. Nucleic Acids Res. 2022 Oct 16:gkac888. DOI: 10.1093/nar/gkac888. PMID: 36243972
Seal RL, Denny P, Bruford EA, Gribkova AK, Landsman D, Marzluff WF, McAndrews M, Panchenko AR, Shaytan AK, Talbert PB. A standardized nomenclature for mammalian histone genes. Epigenetics Chromatin. 2022 Oct 1;15(1):34.DOI:10.1186/s13072-022-00467-2 PMID: 36180920 PMCID: PMC9526256
