Spring newsletter 2025
Newsletters ·
Welcome to James

We are delighted to introduce our new programmer, James McClay, who joined the team in March of this year. James is originally from Devon (UK) and has a BSc (Hons) in Mathematics from the University of Warwick. He is off to a flying start creating a new gene group tool for the curators.
Welcome to plant.genenames.org
Regular readers of our newsletter may recall our introduction to the Plant Gene Nomenclature Committee (PGNC) last summer, our collaboration with the Oak Ridge Center for Bioenergy Innovation. We are excited to announce that the website of this project is now live: plant.genenames.org! The eagle-eyed among you may already have noticed the new tab on genenames.org that takes you through to this website. The PGNC has a consistent look and feel to genenames.org and vertebrate.genenames.org with its own unique background colour 🙂.
We have just published an opinion piece in the journal Tree Physiology (see the Publications section below for a full citation) describing the challenges faced when developing new gene naming guidelines, including feedback from the research community on our draft guidelines for our first plant species, Populus trichocarpa. This is a large deciduous model organism tree commonly known as the black cottonwood. The project already has 439 genes named and we are grateful to our long term collaborator David Nelson for helping us to name the large P. trichocarpa CYP gene family.
Each plant.genenames.org symbol report has a unique PGNC ID and includes the plant species, gene symbol, gene name and gene aliases, as well as links to the relevant NCBI Gene ID, UniProt protein ID and, where available, a PubMed reference ID. One key difference from our other nomenclature websites is the concept of the “primary ID”, which for P. trichocarpa is the Phytozome gene ID (a.k.a the Potri ID) from the Phytozome database. Examples of named genes include AADC1 (aromatic l-amino acid decarboxylase 1) shown in the screenshot below;
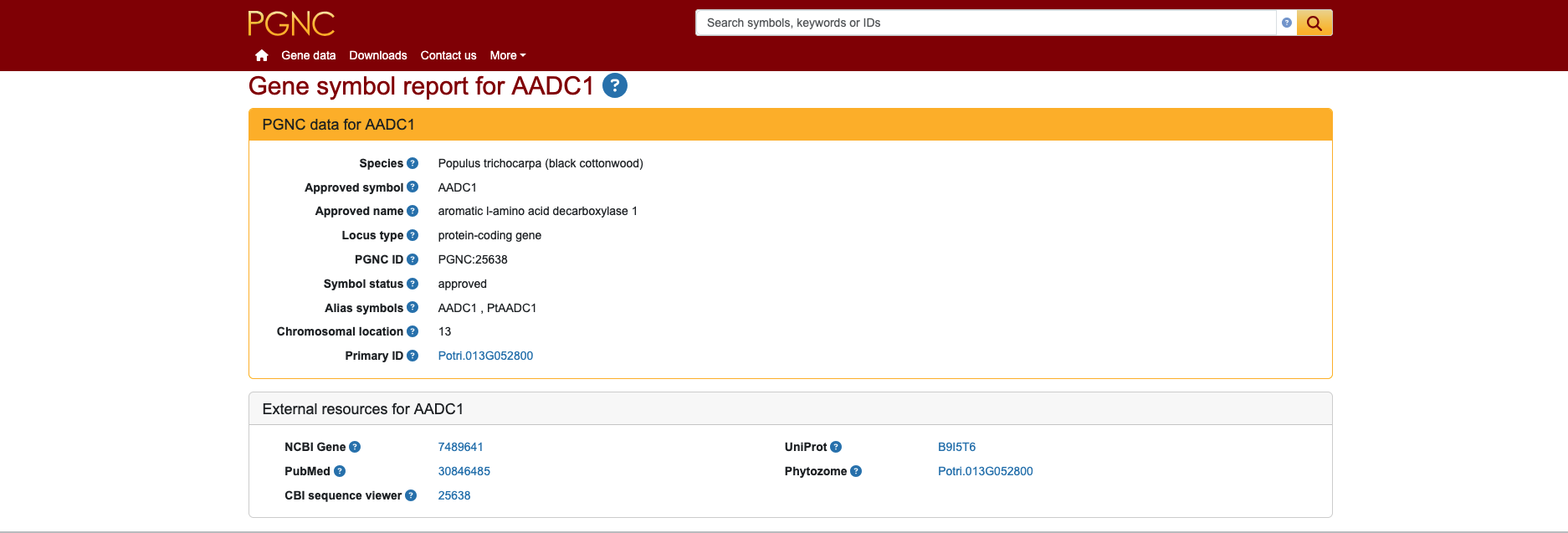
APC1 (anaphase promoting complex subunit 1); BSPA (bark storage protein A); CDC6 (cell division cycle 6). Watch this space for details on another publication that will describe this project in detail!
HGNC gene group news
The HGNC is pleased to announce that we have obtained funding from the SciBite project for Susan to focus on our gene groups for one full day each week. This will allow us to fill in gaps in our gene group resource rather than just adding gene groups as an extra while we name and update genes, which has been our usual practice until now. Susan is beginning this project by focusing on kinase gene groups; we will bring updates on new gene groups in our next newsletter!
Update on genes with the ‘stable’ tag and placeholder symbols
We have added the stable tag to the Symbol Reports of the following genes: ATR, CEP85, CEP85L, CIC, CLMP, CRADD, CREBBP, CXADR, EP300, OTC, PHIP and RNU2-2. This takes our grand total of genes with the stable tag to 3562. Of the latest batch of stabilised genes, no gene symbols were changed, but we modified the names of the following genes to make them clearer or more informative:
- CEP85L > name updated from “centrosomal protein 85 like” to “centrosomal protein 85L”
- CLMP > name updated from “CXADR like membrane protein” to “CXADR like cell adhesion molecule”
- CRADD > name updated from “CASP2 and RIPK1 domain containing adaptor with death domain” to “CARD and death domain containing adaptor protein”
- CREBBP > name updated from “CREB binding protein” to “CREB binding lysine acetyltransferase”
- CXADR > name updated from “CXADR, Ig-like cell adhesion molecule” to “CXADR cell adhesion molecule”
- EP300 > name updated from “E1A binding protein p300” to “EP300 lysine acetyltransferase”
- PHIP > name updated from “pleckstrin homology domain interacting protein” to “PHIP subunit of CUL4-Ring ligase complex”
We have been able to update the nomenclature of two C#orf genes recently: HGNC:29876 has been updated from C1orf43 to LTAP1 for lipid transport auxiliary protein 1 based on data presented in PMID: 40269155. HGNC:28802 has been updated from C18orf21 to RMP24 for ribonuclease MRP subunit p24 based on agreement between groups working on this gene.
Gene Symbols in the News
In a world first, a baby boy has been treated with personalized CRISPR gene therapy created specifically to target his CPS1 disease-causing variant gene. The gene therapy team could not check to what extent the therapy had affected his liver cells as this would have involved a risky biopsy. However, the child is now able to consume more protein and has contracted viral infections without suffering from hyperammonemic crisis, which shows that the therapy has had a positive impact on his health.
A recent study on genes associated with obsessive compulsive disorder (OCD) has identified 30 loci, including the genes WDR6, DALRD3 and CTNND1, which are all candidates for being causative for the disorder. A separate study has identified genetic variations that are candidates for the rarely occurring increased risk of heart problems following COVID-19 mRNA vaccination; variants located near the SCAF11 gene were associated with heart inflammation, while a variant of the LRRC4C gene was associated with risk for myocarditis. Variants of the LRRC4C gene have previously been associated with the susceptibility to and severity of COVID-19.
In every newsletter we like to try to include one of our VGNC species and this time the story is about both a VGNC animal and our own species - the DENND1B gene has been found to be associated with levels of obesity in both dogs and humans. This was a “dog first” discovery - the association was found in labradors, a breed which has higher levels of obesity compared to the average breed, then a search of a GWAS catalog for variants in the orthologous human DENND1B gene found an association with obesity in humans.
Meeting News
Elspeth attended two NIH workshops in early March; “Improving the FAIRness and Sustainability of the NHGRI Resources Ecosystem” and “Data Repositories and Knowledgebase (DRKB) PI Network Meeting” where she got to catch up with colleagues and collaborators from other resources.
Elspeth is also attending ISBT Milan 2025 to take part in an educational session promoting use of standards in genomics.
Publications
Tweedie S, Martin S, Bruford E. Towards an official gene nomenclature for Populus trichocarpa. Tree Physiol. 2025 May 9:tpaf054. doi: 10.1093/treephys/tpaf054. PMID: 40341572
