Introducing the VGNC
VGNC ·
Welcome back to the Genenames blog - this time we’re going to begin discussing our VGNC project, established in 2016 as an extension of the HGNC.
The Vertebrate Gene Nomenclature Committee (VGNC) assigns standardized nomenclature for vertebrate species that lack their own nomenclature committees.
To ensure that genes in model organisms are named in line with their human homologs, we work closely with the five active vertebrate nomenclature committees: MGNC (mouse) , RGNC (rat), CGNC (chicken), XNC (Xenopus) and the ZNC (zebrafish). We will be discussing how we work with the model organism databases (MODs) further in a later blog post.
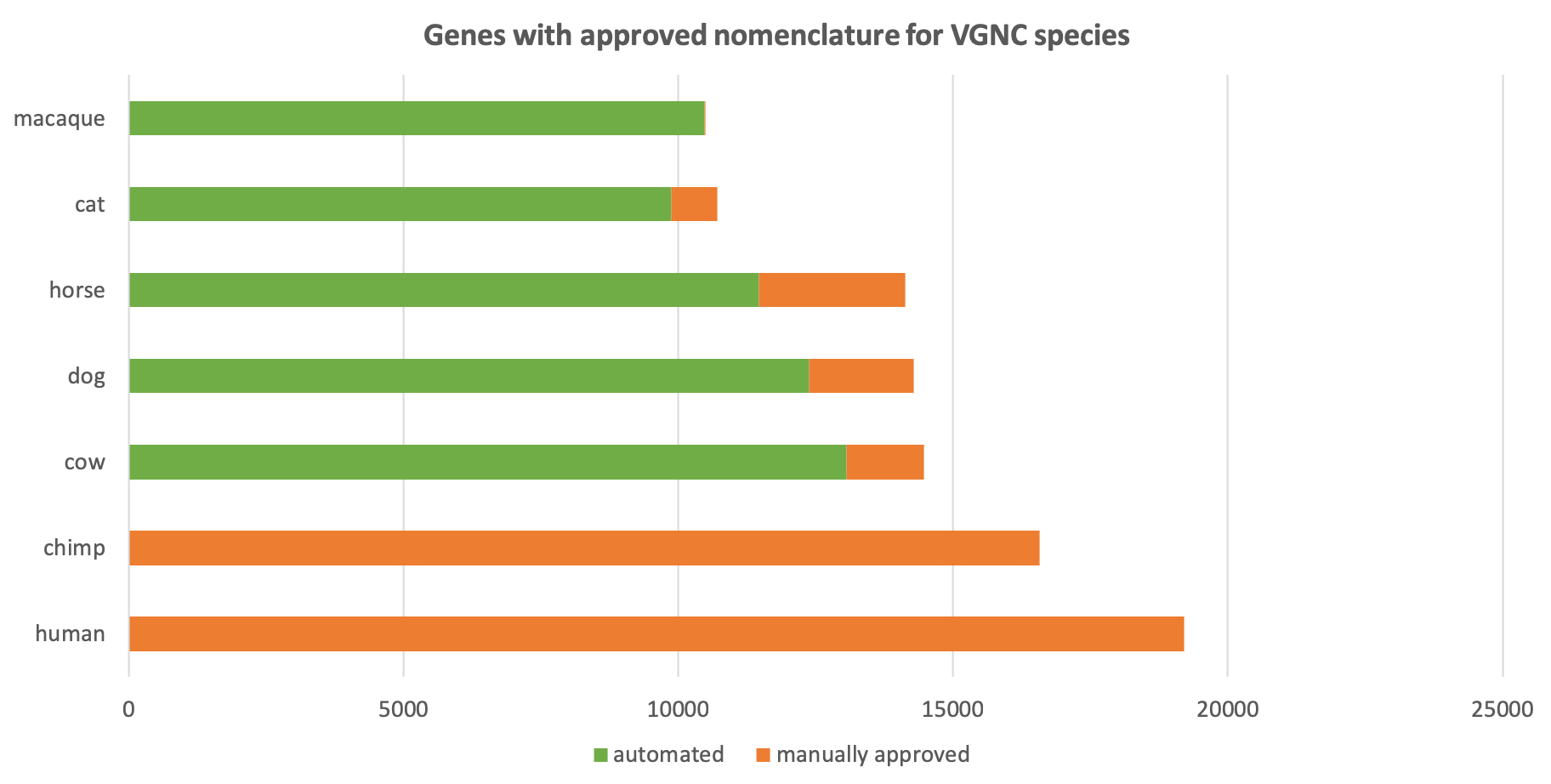
We started the VGNC project in 2016 with the chimpanzee (Pan troglodytes) as our pilot project species: chimps are the closest living evolutionary relatives to human and hence their genome is relatively similar to ours .
Although chimp is no longer heavily used as a biomedical model (and indeed, the use of great apes in biomedical research has been banned by several countries), its genomic comparison with human may still provide insights that can be useful in the study of human disease.
We have since expanded our VGNC naming to horse, cow and dog and our two latest additions, cat and macaque. We’ll update you here as we add new species to the list!
The VGNC uses a software pipeline based on the HGNC Comparison of Orthology Predictions (HCOP) tool to identify equivalent genes in other species. We use data from four different resources: Ensembl, NCBI Gene, OMA and Panther and where they all agree, the equivalent gene is automatically given the same name as the human gene.
For genes where there is less agreement among these resources, naming is based on a combination of a semi-automated pipeline and manual curation, taking into account other data such as gene order (synteny) and sequence similarity (by phylogeny).
We also rely on the expertise of specialist advisors for naming within complex gene families, where there have been lots of gene gains and losses throughout evolution (for example, the olfactory receptor gene family).
As our pilot project, we manually curated the naming of all of the chimp genes: this has enabled us to review our human gene nomenclature, ensuring that it is appropriate to transfer across vertebrates. We are keen to avoid reference to human traits and diseases in gene nomenclature wherever possible, as they may not be relevant across species.
All VGNC data is available via our dedicated website https://vertebrate.genenames.org and the NCBI Gene and Ensembl databases now display approved VGNC nomenclature for all relevant entries, along with reciprocal links to the symbol reports on the VGNC site.
Have you ever wondered about the donors selected to have their genomes sequenced as representatives of their species? The Human Genome Project (HGP) recruited a large number of individuals and only a few of their samples were sequenced to create the human genome, with donor identities remaining anonymous.
However, for the current vertebrate species included in VGNC, most of the DNA sequenced has come from individual animals. We can therefore acknowledge Clint (chimpanzee genome project), Tasha (dog genome project), Twilight (horse genome project), L1 Dominette 01449 (cow genome project) Cinnamon (cat genome project) and ID 17573 (rhesus macaque genome project) for their contributions to genomics.
Is there a particular species you would like to see included in the VGNC resource? Please email us if you have expertise in a particular species or gene family you could help us to name: vgnc@genenames.org.
