HCOP: the HGNC Comparison of Orthology Predictions tool
HGNC, HCOP ·
While most of you will mainly be interested in human genes, did you know that we also provide an easy way to find orthologs (equivalent genes in another species) of human genes using our HCOP (HGNC Comparison of Orthology Predictions) tool?
HCOP brings together data for 19 other species, from a range of different orthology resources, and provides a quick visual comparison of all the data. The resources we use are EggNOG, Ensembl, HGNC, HomoloGene, Inparanoid, OMA, OrthoDB, OrthoMCL, NCBI Orthologs, Panther, PomBase, PhylomeDB, Treefam and ZFIN. The more resources that agree on an ortholog, the greater the confidence you can have in that ortholog being correct.
As well as ortholog data we also collect gene nomenclature data daily from model organism databases (MODs - MGI, RGD, Xenbase, ZFIN, WormBase, SGD and PomBase), to ensure that we are showing you the latest gene symbols and names for each species. This also means that if you search HCOP using a symbol from a MOD you should be able to find a result.
To identify orthologs of your gene of interest you can search HCOP using either an Ensembl gene identifier or an NCBI Gene identifier. If the gene belongs to a species with a MOD or nomenclature database you can also search with the approved gene symbol, approved gene name or the gene identifier from that database. You can also query HCOP with more than one gene at a time - either by listing the search terms in the ‘enter identifier(s)’ box or by uploading a list of search terms as a file using the ‘upload file’ option. If you only want the HCOP results to show results for human and a specific set of species, or to only include results from a particular ortholog resource (or resources), just tick the checkboxes on the search form.
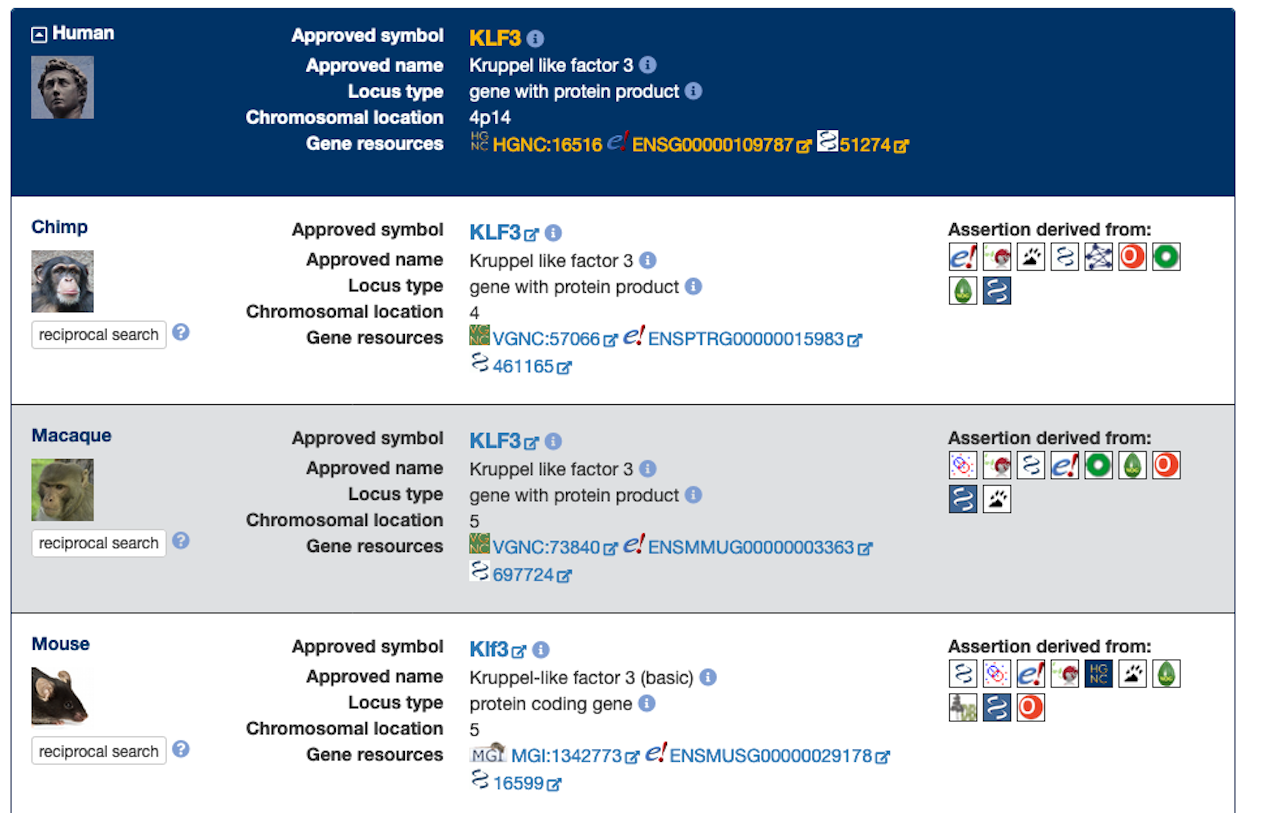
If your query brings back one or more results within HCOP you will see a panel like the one shown below. The dark blue section contains information about your query gene, and each of the lower white or light blue sections represents an orthology assertion for that gene. Each assertion section contains the following information: the species that the ortholog is from; summary data about the ortholog, including links to databases such as NCBI Gene and Ensembl; and, most importantly, the resources that support that assertion. The higher the number of assertions the more confidence you can have that this is a genuine ortholog. If there are multiple assertions returned for any species, the assertion supported by the most resources is shown first.
We also have pre-computed some files of HCOP data that you can download using the bulk downloads tool on the HCOP pag or from our FTP site. You can choose a file containing human ortholog data for a single species, or human ortholog data from all 19 HCOP species in a single file.
If you would like to see us include orthology data from an additional source, or would like us to add a new species to HCOP, please just let us know via hgnc@genenames.org
