New Guidelines 2020
HGNC ·
A closer look at our more recent HGNC guideline updates
The last blog post featured our Summer Newsletter, and we discussed some of the issues brought up in our Nature Genetics comment article on our guidelines including mention of our solution for the “Excel auto-changing date symbols” problem. A summary of our current guidelines can be found on our website.
We hope you have had the time to read our paper and thought you might find it interesting to read a little more about some of the updates we have made to our guidelines, with some extra examples that we didn’t have room to mention!
Naming may be based on recognized structural domains and motifs encoded by the gene
If the function of a gene is unknown, but its encoded protein contains a particular domain and/or motif, this can give an insight into its likely function. Gene symbols assigned based on domains are often retained with extra information added to the gene name once the function of its protein product has been determined.
For example:
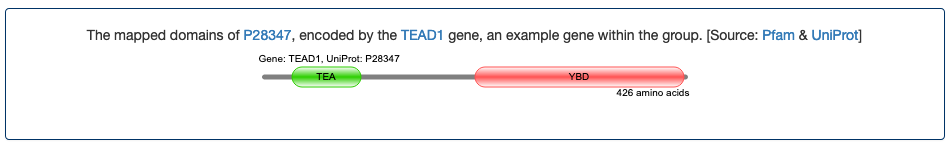
- TEAD1 (TEA domain transcription factor 1). The TEA domain (also known as the ATTS domain) has been characterized as a DNA binding region. This domain based naming provided information about the likely function of this set of genes encoding TEA domain containing proteins before they were shown to encode transcription factors.
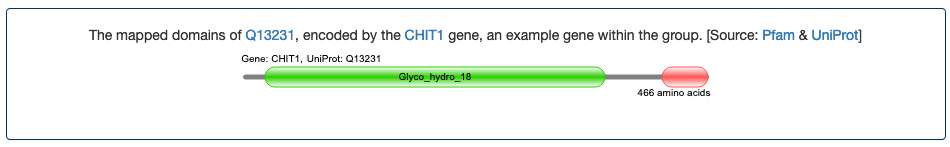
- CHID1 (chitinase domain containing 1). This gene encodes a protein containing a chitinase domain and is a member of the chitinase family. The chitinases are hydrolytic enzymes that break down glycosidic bonds in chitin. Despite its evolutionary relationship with the chitinases, CHID1 has been published as encoding a protein with cytokine-like and growth factor-like properties but no enzymatic activity.
Naming may be based on homologous genes within the human genome
A shared root symbol can be used to reflect paralogy, with each member receiving the next number or letter in the series. Genes of unknown function in an established family are given the next symbol in the series but with a different gene-name format. If there is no functional information known about any family members then all genes in that family may be given a placeholder FAM# root. This may be updated at a later date, or retained if it has become established in the literature and researchers are keen to do this.
-
FAM20A (FAM20A golgi associated secretory pathway pseudokinase), FAM20B (FAM20B glycosaminoglycan xylosylkinase) and FAM20C (FAM20C golgi associated secretory pathway kinase). We contacted all authors who have previously published on these genes to propose a possible nomenclature update. The majority of authors publishing on these genes wanted to retain the well published FAM20# root symbol, while tweaking the associated gene names to make them more informative.
-
ABI1 (abl interactor 1), ABI2 (abl interactor 2) and ABI3 (ABI family member 3). In this case the ABI3 protein has not been reported to interact with the ABL1 (ABL proto-oncogene 1, non-receptor tyrosine kinase) protein product.
Naming may be based on homologous genes from another species
Some human genes are named based on homologous characterized genes from another species. In the past, the name of the species that we based this on would have also been included in the human gene name in brackets, but many users told us this caused confusion. Furthermore, now that we name genes in other vertebrate species, we can appreciate that sometimes including reference to a specific species in a gene name can be confusing. We have now removed these references from our gene names.
When naming is based on a 1:1 orthology relationship between human and another species we use the same, or at least an equivalent, symbol as used in the species in question.
For example:
- RAE1 (ribonucleic acid export 1), previously “RAE1 RNA export 1 homolog (S. pombe)” is also called rae1 in S.pombe.
- EME1 (essential meiotic structure-specific endonuclease 1), previously “essential meiotic endonuclease 1 homolog 1 (S. pombe)” is also called eme1 in S.pombe.

- SUPT3H (SPT3 homolog, SAGA and STAGA complex component) was named based on its homology with the S.cerevisiae SPT3 gene. We could not use the symbol SPT3 in human, as this clashed with the root symbol SPT# already approved for a group of unrelated genes, the spectrins. The word “homolog” was kept in the gene name to maintain the link with yeast nomenclature.
- BUD23 (BUD23 rRNA methyltransferase and ribosome maturation factor). This was named after the S.cerevisiae ortholog BUD23 and the symbol was updated from the phenotype associated WBSCR22 (Williams Beuren syndrome chromosome region 22).
A unique number or letter suffix is added if there is more than one human homolog:
- SEC22A (SEC22 homolog A, vesicle trafficking protein), previously called “SEC22 vesicle trafficking protein homolog A (S. cerevisiae)”, SEC22B (SEC22 homolog B, vesicle trafficking protein), previously called “SEC22 vesicle trafficking protein homolog B (S. cerevisiae)” and SEC22C (SEC22 homolog C, vesicle trafficking protein) previously called “SEC22 vesicle trafficking protein homolog C (S. cerevisiae)”.
The single yeast ortholog of these genes is called SEC22 in the Saccharomyces genome database (SGD).
Some gene nomenclature needed to be updated to make it more appropriate and/or informative for use in human. For example:

-
BICD1 (BICD cargo adaptor 1) was previously named “Bicaudal D (Drosophila) homolog 1”
-
DLGAP1 (DLG associated protein 1) was previously named “discs large homolog 5”
-
ASXL1 (ASXL transcriptional regulator 1) was previously named “additional sex combs like 1 (Drosophila)”

- AKTIP (AKT interacting protein) was previously assigned the symbol FTS alongside the name “fused toes (mouse) homolog)”
Finally, some gene names ported over from homologs in other species needed to be changed because as well as being inappropriate in human they could also be perceived as offensive or pejorative.
For example:
- HECA (hdc homolog, cell cycle regulator) was previously named “headcase homolog (Drosophila)”.
Naming may be based only on the presence of an open reading frame
If there is an open reading frame but nothing is known about the gene product, it can be assigned a C#orf# placeholder symbol. The numbering is designated by chromosome, the letters “orf” standing for “open reading frame” and an iterative number. KIAA# is another placeholder symbol that has been approved for genes identified by the Kazusa cDNA sequencing project.
You can read more about placeholder symols in this previous blog post.
We no longer name…
HGNC historically named human loci associated with clinical phenotypes. This role has been transferred to OMIM, and as our existing phenotype records were not complete or being regularly updated we decided to withdraw them. Hence all previous “phenotype only” HGNC records now have the status of ‘entry withdrawn’. However, some phenotype based gene symbols have been highly published and so have been retained, with updates to their associated gene names to reflect the normal function of the proteins they encode.
You can read our previous blog posts to find out why we are now striving to stabilize gene symbols and about why nevertheless, we sometimes do make changes to gene symbols when there are compelling reasons to do so.
Comments or questions about our guidelines?
If you’ve enjoyed reading our comment article on our guidelines but have further specific questions about gene naming, you can contact us via email at hgnc@genenames.org.
