Naming genes in the mitochondrial genome
HGNC ·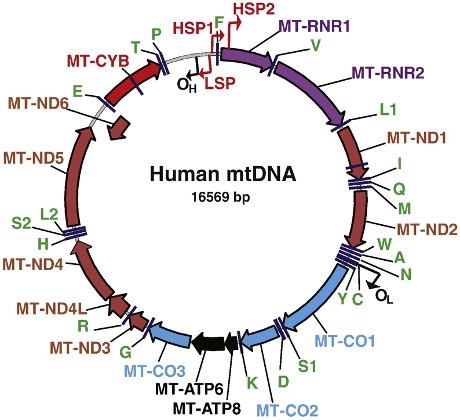
Figure reproduced from Shololenko and Alexeyev, 2015 with permission from M. Alexeyev (PMID: 26071375).
Mitochondria are vital for cellular energy and these mini power stations require the coordinated effort of over 1000 proteins (PMID:20690818). While the majority of these are encoded by genes within the nuclear genome, many having moved there from the mitochondrial genome in the course of evolution, 37 of them remain on the mitochondrial genome. While, depending on the conditions, the number of mitochondria in cells may vary dramatically you certainly can’t live without them completely (PMID:26071375) and, given the key function of the mitochondrion, it isn’t surprising that mutations in genes for mitochondrial proteins are often associated with disease. Over 350 genes have already been associated with mitochondrial diseases resulting in a diverse array of problems affecting all parts of the body particularly the nervous system and muscles (PMID:34146515). One example is ‘myoclonic epilepsy with ragged red fibres’ (MERRF syndrome) - around 90% of cases of MERRF syndrome are due to single nucleotide change in the mitochondrial lysine tRNA gene (MT-TK). The study of mitochondrial disease is a hot topic - it will be interesting to see if work suggesting the CRISPR/Cas9 System can be used to target the mitochondrial genome (PMID:26448933, PMID:33889176) can be replicated and open a potential new route to treating these disorders. Clearly it is important that we name all genes relevant to mitochondrial function and don’t forget the ones on the mitochondrial genome itself.
The vast majority of the mitochondrially encoded genes were given approved nomenclature around thirty years ago (PMID:1463005) and they are immediately recognizable because their symbols all begin ‘MT-‘ (e.g. MT-ATP6). In general, we do not use punctuation in symbols but the inclusion of a hyphen in these symbols is one of the few special exceptions. A similar notation is used in the mouse but with lower case mt (e.g. mt-Atp6). As the ‘MT-’ is often dropped by authors, particularly when mitochondrial genes are the only genes under discussion, we record ‘MT-’ free symbols such as ATP6 as aliases.
The gene names also make it clear that these genes are encoded by the mitochondrial genome (e.g. ‘mitochondrially encoded ATP synthase membrane subunit 6’) to distinguish these from nuclear encoded genes named for mitochondrial function e.g. MTCH1 (mitochondrial carrier 1). Nuclear genes that include ‘mitochondrial’ in their name typically include MT in their gene symbol though in some cases this may simply be M e.g. MECR (mitochondrial trans-2-enoyl-CoA reductase).
You can see the full set of records with the ‘MT-’ root symbol via our gene groups e.g. search for ‘mitochondrial’ and filter for groups - the top hit is ‘Mitochondrial genome’.
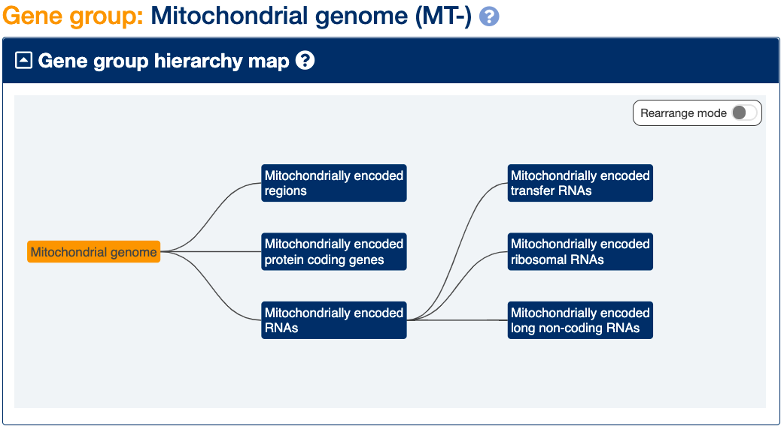
The 37 highly conserved genes in the mitochondrial genome comprise 13 protein coding genes, 22 transfer RNAs and 2 ribosomal RNAs. The protein products are all components of the mitochondrial oxidative phosphorylation system. You will notice that, for historic reasons, the mitochondrially encoded tRNAs are named in a simpler format - using the single letter amino acid code (e.g MT-TR for “mitochondrially encoded tRNA-Arg (CGN)”) - to the tRNAs encoded by the nuclear genome which include the anti-codon in the symbols (e.g TRR-ACG1-1 “tRNA-Arg (anticodon ACG) 1-1”). The tRNAs encoded by the nuclear genome that are used for mitochondrial translation have their own root symbol NMTR (e.g. NMTRQ-CTG1-1 “nuclear-encoded mitochondrial tRNA-Gln (CTG) 1-1)” to distinguish them from tRNAs used for translation in the cytoplasm.
Beyond the standard MT products, we have also named a single long non-coding RNA MT-LIPCAR which was identified as a biomarker for cardiac remodeling (PMID:24663402). However, there is some doubt as to whether this transcript arises from mitochondria or mitochondrial DNA integrated into the nuclear genome (PMID:24812345). There is also evidence that a 24 residue peptide is produced from the mitochondrial 16S ribosomal RNA (MT-RNR2) - this peptide, called humanin, is reported to be protective against Alzheimer disease related neurotoxicity (PMID:12009529). At present, we record ‘humanin’ as an alias of MT-RNR2 rather than naming it as a separate protein coding gene.
You can see we also have a group called ‘Mitochondrially encoded regions’ - these cover key non-coding regions of the mitochondrial genome including the origins of replication (MT-OLR and MT-OHR) and promoters (MT-HSP1, MT-HSP2, MT-LSP) for the light and heavy strands respectively. Naming this type of feature is feasible for the relatively small and well defined mitochondrial genome but is currently beyond the scope of what we can name in the nuclear genome. Please let us know if you think we should be naming more of these elements. All of these regions are included in the MITOMAP resource (MITOMAP: A Human Mitochondrial Genome Database, 2019), which provides extensive information about mitochondria including details of mitochondrial alleles and associated references. We provide links to MITOMAP from our gene group pages.
Downloading the MT genes
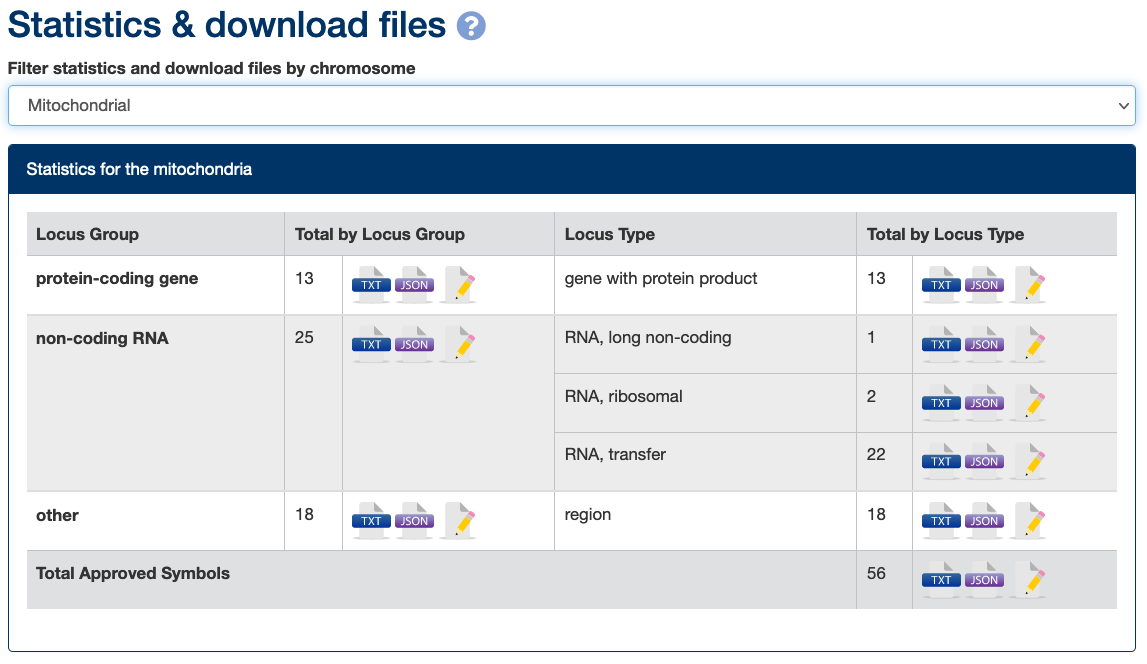
It is easy to download our data for genes on the mitochondrial genome (or subsets of it) via the gene groups pages or from our statistics and downloads page which includes the option to filter by chromosome.
Pseudogenes of mitochondrially encoded genes
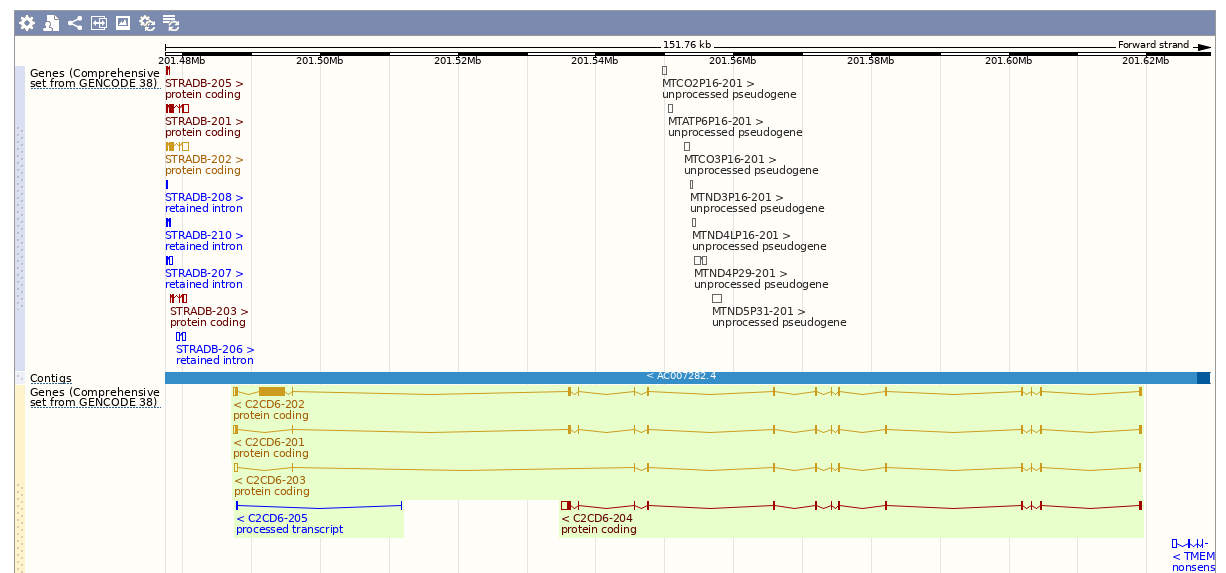
Over the course of evolution, chunks of mitochondrial DNA are occasionally incorporated into the nuclear genome giving rise to nuclear mitochondrial pseudogenes termed NUMTs. These pesky pseudogenes, which may be very similar to true mitochondrial genes, can cause problems in disease studies by misleading researchers into believing they have found a new MT gene variant (PMID:18611982). We name these pseudogenes after the parental mitochondrial gene in a numerical series but, importantly, their symbols do not contain a hyphen as they are not part of the actual mitochondrial genome. For example, we have named 57 MT-CO1 pseudogenes: MTCO1P1 to MTCO1P57 - the gene names are simply ‘MT-CO1 pseudogene 1’ etc, referencing the parent gene but avoiding any suggestion that they are on the mitochondrial genome. These genes appear in small clusters reflecting the gene order in the parent genome - the example below shows a NUMT cluster located in a C2CD6 intron. Note that there is no significance to the numbering of these pseudogenes - they were simply named (and numbered) as they came to our attention.
Many thanks to Mikhail Alexeyev for allowing us to reproduce the figure of the mitochondrial genome originally published here: (https://www.sciencedirect.com/science/article/pii/S0925443915001696). The ribosomal RNA and protein-coding genes are labelled with HGNC approved gene symbols; tRNAs are represented by the single letter representation of the amino acid they code for.
