Summer newsletter 2021
Newsletters ·
Dinas Dinlle beach, Wales
Welcome to Liora!
We are delighted to introduce our new curator, Liora Vilmovsky, who has joined us to work primarily on our project to stabilise symbols for clinically relevant genes. You can read Liora’s post on our blog site to learn more about her. We are looking forward to meeting her in person when the group begins to return to the office, hopefully in the autumn.
Symbol change notification files
We are about to make FTP files available in our HGNC archive that will show a list of symbol changes for the span of a month or a quarter of a year. The file will contain the HGNC ID, approved symbol, previous symbol, gene name, locus type, and the date the symbol changed. The files will be created on the 1st of each month and quarterly. The first monthly file will be available on September 1st and the first quarterly file will be available on October 1st. Users can sign up for notification emails that will alert them to the presence of a new FTP file via our feedback form, which now contains the checkbox “Receive symbol changes notification email”.
Genes with the ‘stable’ tag
As mentioned above, Liora has joined the team to work on our symbol stabilising project. She and other curators are in the process of manually reviewing clinically relevant genes and marking them with the tag ‘stable’ if we think it is extremely unlikely that we will ever change the gene symbol. Genes that we have marked as stable display the blue ‘stable symbol’ tag  at the top of the Symbol Report, see MTOR for an example. You can view the number of genes that we have marked as stable using our ‘Filter by stable symbol tag’. Simply press return in our search box and then perform the stable filter; as of August 16th 2021 we have 2443 stable symbols.
at the top of the Symbol Report, see MTOR for an example. You can view the number of genes that we have marked as stable using our ‘Filter by stable symbol tag’. Simply press return in our search box and then perform the stable filter; as of August 16th 2021 we have 2443 stable symbols.
New gene groups
We have recently curated further methyltransferase gene groups and placed these into our Methyltransferases gene group hierarchy.
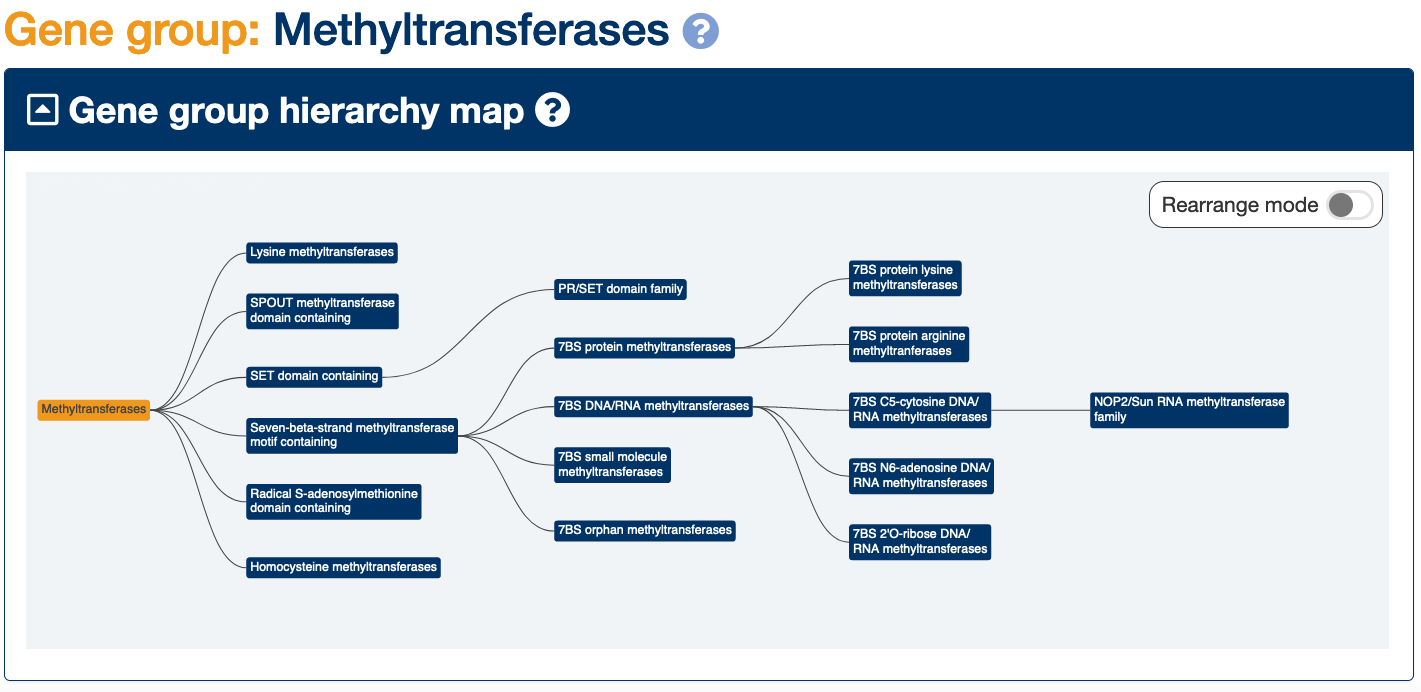
The Methyltransferases group at the top of the hierarchy now contains two additional curated subgroups: the Homocysteine methyltransferases and the Seven-beta-strand methyltransferase motif containing group, as well as four pre-existing subgroups.
The Seven-beta-strand methyltransferase motif containing group contains the 7BS small molecule methyltransferases, the 7BS orphan methyltransferases, the 7BS protein methyltransferases and the 7BS DNA/RNA methyltransferases as subgroups.
The 7BS protein methyltransferases are then split into two subgroups: the 7BS protein arginine methyltranferases and the 7BS protein lysine methyltransferases.
The 7BS DNA/RNA methyltransferases can be sorted into three subgroups: the 7BS C5-cytosine DNA/RNA methyltransferases,the 7BS N6-adenosine DNA/RNA methyltransferases and the 7BS 2’O-ribose DNA/RNA methyltransferases.
You may find the ‘Gene group hierarchy’ maps at the top of each methyltransferase gene group page a useful way of navigating through this complex hierarchy!
Gene Symbols in the News
We start this section with two new associations reported between human genes and disease: A rare loss of function SUV39H2 variant gene was identified in a person with an autism spectrum disorder (ASD), while reduced expression of SUV39H2 was found in post mortem brains of people with ASD. Mutations were discovered in the PNLDC1 genes of four unrelated individuals with non-obstructive azoospermia. The PNLDC1 gene encodes an RNA exonuclease that processes the 3’ end of piRNA genes, non-coding RNAs that are necessary for spermatogenesis.
In a bid to understand why females live on average longer than males and with a lower degree of neurodegeneration, a group began studying the KDM6A gene, which escapes X inactivation in brain cells, meaning that females express higher levels of this gene. They found that XX brain cells were more resistant to toxins, including beta-amyloid.
A recent study has found that the ELOF1 gene encodes a multifunctional protein that is a transcription elongation factor which also takes part in transcription-coupled nucleotide excision repair and regulates transcription to protect cells from transcription-mediated replication stress. This gene had not been identified in previous screens of cells from patients with DNA repair defects because ELOF1 is an essential gene and so was not thought to be linked to disease.
In COVID-19 related news, a study on the genomes of patients with a severe form of the disease has found variants of the following genes are associated with increased risk of severe disease:IFNAR1,IFNAR2, FOXP4, DPP9 and genes of the OAS family. Although these variants only increase risk by a relatively small amount, they may provide information on the mechanism of the disease.
A separate study found that carriers of the HLA-DRB1*04:01 allele of the HLA-DRB1 gene are three times more likely to have asymptomatic COVID-19 infections than the general population. Finally, activation of the IFI27 gene has been found to be able to predict COVID-19 infections up to a week before patients test positive for the disease - caution is needed as this gene is also activated by other viruses, but this knowledge could be useful in predicting cases in COVID-19 contacts ahead of later positive PCR tests.
We end with news about a horse gene! A recent study has provided extra insight on a mutation of the STX17 gene that causes horses to go grey with age. This dominant mutation was thought to be due to a 4.6kb duplication within an intron of the STX17 gene, but recent droplet digital PCR work has shown that the causative region actually contains a triplication of the 4.6kb segment.
Meeting News
Tamsin attended the Quest for Orthologs 6.5 virtual conference on 2nd-3rd August. She chaired a session and gave a talk entitled “Updates to HCOP: the HGNC comparison of orthology predictions tool”, which was at 6:30am British Summer Time!
