Stability in the time of COVID-19
HGNC ·
Coronavirus disease 2019 (COVID-19), an ongoing pandemic caused by a new form of coronavirus known as SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), has dominated the news for the past 18 months and still influences our lives in many aspects.
International research on COVID-19 and SARS-CoV-2 molecular biology is well underway with government organisations, academic universities and institutes and industry researchers all being heavily involved. The COVID-19 UniProtKB webpage publishes the latest pre-release UniProtKB data for the SARS-CoV-2 proteome and other proteins related to the COVID-19 outbreak. To date, the site reports 112 proteins that have been identified as being associated with COVID-19, 81 of which are encoded in the human genome. This group of genes include immune related genes, transcription factors, enzymes and receptors.
As part of our ongoing efforts to stabilize gene symbols with an emphasis on genes associated with human diseases, we decided to prioritize reviewing the nomenclature of genes linked to COVID-19. We have also noticed a great increase in the research and publication of manuscripts discussing and investigating some of these genes. TMPRSS2 (transmembrane serine protease 2) is one example of a gene that has recently been increasingly referenced in publications (see Figure 1), with an average of 642.5 papers for 2020-2021, compared to 56.8 publications for the years 1997-2019. The use of HGNC’s standardised and informative gene symbols can help researchers identify with certainty which genes are being discussed in these papers. This helps prevent confusion that could otherwise result from this flood of SARS-CoV-2 related publications.
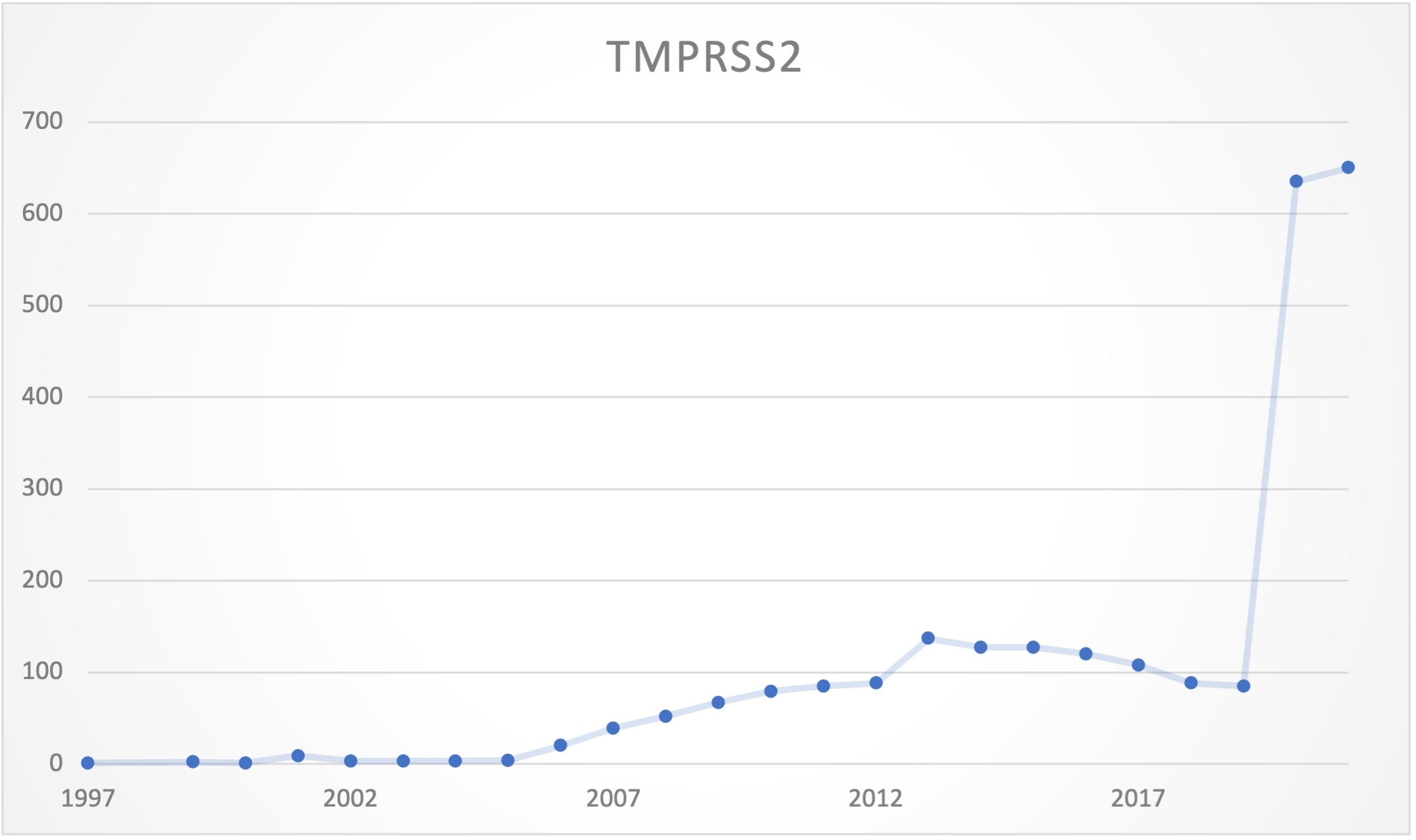
Figure 1. TMPRSS2 number of publications per year.
We have now examined the nomenclature of all 81 genes related to COVID-19 in humans, and have stabilized 83% of their gene symbols. When we look to stabilize a gene symbol (essentially saying that it is very unlikely to be changed in the future), our primary aims are to ensure the basis for naming the gene is still true and there is nothing that could be construed as offensive in the name or symbol. We also look for anything that is causing confusion and try to resolve it– sometimes a symbol is also used as an alias to refer to a completely different gene. If this is a significant issue then a change to the approved symbol may perhaps be appropriate. For genes that have temporary symbols or relatively uninformative names, we see if we can update them to something more meaningful. We normally prefer to name a gene in relation to its function, or functional motif/domain and not for any disease or syndrome it might cause or relate to. We examine aliases for the gene, and check whether any aliases are missing, and whether an alias is more appropriate to be the approved symbol. Finally, we verify that the information and links on our records are correct, complete and up to date.
There are still 14 COVID associated genes left that have not yet been stabilized for various reasons; some might be easy to resolve and others may be more problematic. For example, we have not stabilized the symbol for transmembrane protein 41B (TMEM41B), because the gene name is still comparatively uninformative. This nomenclature has not yet been widely used (30 publications on PubMed), and therefore, the effect of a symbol change could be minimal. We will investigate renaming TMEM41B, and potentially its paralogs TMEM41A and TMEM64, based on the recent publications describing the function of TMEM41B as a phospholipid scramblase.
In some cases we have found that the alias of a gene is used more than the approved symbol, even after the symbol was officially approved. For example, NKG2A and CD94 are the aliases of two of the killer cell lectin-like receptors, KLRC1 and KLRD1, and are used more often than the approved symbols. A search for CD94 in PubMed results in 1011 papers, while only 374 publications appear for KLRD1. The use of aliases instead of approved symbols can cause readers to miss relevant papers relating to their gene of interest. We have decided to review the approved symbols and aliases in the killer cell lectin like receptor group and consult with experts before stabilising any of these symbols.
NFE2L2, nuclear factor, erythroid 2 like 2, is another example where the alias, NRF2 is more popular than the approved symbol. However, NRF1 is already an approved gene symbol for ‘nuclear respiratory factor 1’ and therefore, the use of NRF2 for NFE2L2 would be likely to cause confusion. We have decided to retain NFE2L2 as the approved symbol, which also keeps it consistent with the other NFE2-like family members (NFE2L1 and NFE2L3).
Following our review of these genes, the only gene symbol that we plan to change so far is PHB which encodes one of two prohibitin genes in the human genome. From our review, it is clear that, in contrast to the second prohibitin gene symbol PHB2, “PHB” is a very poor search term for retrieving papers about this gene. “PHB” is an abbreviation for many other things, including poly-beta-hydroxybutyrate which accounts for almost 2000 ‘PHB’ publications. Within papers about prohibitin, PHB is used variously to refer to this specific PHB gene, the prohibitin family, the prohibitin homology domain and the prohibitin complex.
However, the alias “PHB1” has already been used in many papers to reference the specific human gene/gene product and its vertebrate orthologs, and its use has increased over time. We believe changing the approved gene symbol from PHB to PHB1 would make it easier to refer to the gene specifically, thereby improving communication and, in the longer term, would help researchers find relevant literature . This update would also bring the vertebrate nomenclature into line with orthologs in other species (e.g. C. elegans (phb-1), S. cerevisiae (PHB1) and S. pombe (phb1) where both paralogs are numbered. We have written to key researchers working on PHB to advise them the symbol will change in October and to date have received only supportive feedback.
We aim to resolve the various issues for the remaining genes and stabilize all gene symbols currently reported as relating to COVID-19 in time. Now we only need to ensure we have the cooperation of scientists and scientific journals to use only approved gene symbols in all publications!
