The risks of using unapproved gene symbols
HGNC ·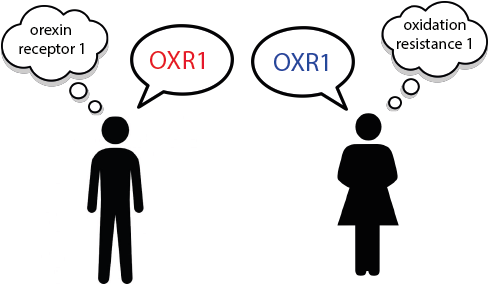
Does it really matter if researchers choose to use their favourite aliases to refer to genes in their manuscripts instead of using their HGNC approved symbols? Sadly, the use of unapproved aliases can not only cause confusion and wasted experiments in the laboratory, but even more worryingly, confusion in the clinic has the potential to cause harm to patients.
There can be little doubt that researchers can discuss their work with the wider scientific community more easily when they use unique, approved, and functionally informative nomenclature in their publications.
You may have already read our recent commentary paper “The risks of using unapproved gene symbols” (October 2021) published in AJHG. In this paper we discuss several examples of cases where genes have been confused due to either the use of shared alias symbols or because an approved symbol clashes with an alias.
In case you haven’t had a chance to read it yet, here are two examples…
TAFAZZIN and WWTR1
For our first example, until very recently the approved symbol for the gene encoding the protein “tafazzin” was TAZ (HGNC: 11577; MIM: 300394). This gene is of clinical interest because variants are associated with the X-linked genetic disorder Barth syndrome (MIM: 302060). The tafazzin protein plays a key role in cardiolipin remodeling and so is vital to mitochondrial function. Barth syndrome patients typically have a form of cardiomyopathy associated with an enlarged heart, along with skeletal muscle myopathy and neutropenia, which can result in a weakened immune system (PMID: 33001359).
Unfortunately, the gene approved as WWTR1 (WW domain containing transcription regulator 1) (HGNC: 24042; MIM: 607392) has been heavily published with the alias symbol “TAZ,” alongside the name “transcriptional coactivator with PDZ-binding motif.” The WWTR1 protein interacts with the protein encoded by YAP1 (Yes1 associated transcriptional regulator) (HGNC: 16262; MIM: 606608), and the two are often referred to together as “YAP/TAZ”: a pair of transcriptional co-activators of the Hippo signaling pathway (PMID: 33997171).
This situation not only makes the retrieval of relevant papers about tafazzin and the protein encoded by WWTR1 more difficult but can also lead to confusion between the two. For example, Takehara et al. (2018) (PMID: 29731861) had to be retracted by its authors. This group was studying the Hippo pathway and its role in the development of mesothelioma. They intended to use the WWTR1-encoded protein in their experiments, but because of the common usage of the alias “TAZ,” they confused their gene of interest with tafazzin and used the incorrect protein as part of their studies.
The HGNC discussed whether we could make a change to the gene nomenclature to help avoid the recurrence of this situation. Although the researchers working on the approved TAZ (tafazzin) gene were correctly using the approved nomenclature, we decided that a pragmatic solution to minimize further confusion could be to update the approved symbol for TAZ (HGNC: 11577) to TAFAZZIN in line with the name of the encoded protein, providing that the community working on this gene were supportive of this change. The vast majority of researchers working on this gene already used the protein name “tafazzin” in their publications.
We wrote to authors who had previously published on TAZ (tafazzin) and, fortunately, most could see the benefits of this proposal and were in favor of it. In addition, the Barth Syndrome Foundation wrote us a letter of support, reassuring us that the clinical community felt that this would be a justified symbol change to avoid confusion and that they would use the new nomenclature once updated.
A few researchers understandably felt that the “YAP/TAZ” community should just stop using the symbol “TAZ” for the WWTR1 gene. However, unfortunately we cannot “police” the use of aliases, although we can advocate to journals and authors about how important it is to use standardized, approved nomenclature when discussing genetics.
Our March 2021 symbol update of HGNC: 11577 to the approved symbol TAFAZZIN (tafazzin, phospholipid-lysophospholipid transacylase) should help minimize future confusion and aid data retrieval by allowing papers about the tafazzin gene to be more easily separated from those discussing the “YAP/TAZ” pathway.
OXR1 and HCRTR1
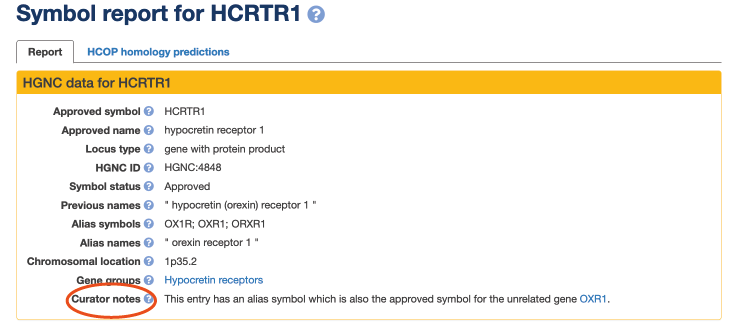
A researcher recently wrote to us (April 2021) expressing concern over the confusion in the literature caused by the publishing of the symbol “OXR1” as an alias for the gene approved as HCRTR1 (hypocretin receptor 1) (HGNC: 4848; MIM: 602392).
The symbol OXR1 is currently approved for the “oxidation resistance 1” gene (HGNC: 15822; MIM: 605609), which has been associated with a human phenotype involving cerebellar hypoplasia/atrophy, epilepsy, and global developmental delay ((MIM: 213000)). As its encoded protein protects neurons from oxidative stress-induced apoptosis, it is also being investigated in terms of possible therapeutic effects in the treatment of amyotrophic lateral sclerosis (MIM: 105400).
The unrelated genes HCRTR1 and HCRTR2 (HGNC: 4849; MIM: 602393) encode proteins that act as receptors for the neuropeptide hormones known as the hypocretins, also published as “orexins.” A variant of HCRTR2 has been associated with narcolepsy in dogs (PMID: 10458611) and humans (PMID: 10615891) and “orexin” knockout mice also show a phenotype similar to human narcolepsy (MIM: 161400) (PMID: 10615891).
The symbol aliases “OX1R” and “OX2R” have previously been used to refer to HCRTR1 and HCRTR2, (PMID: 25957175) but the letter order of the “OX1R” alias symbol for HCRTR1 was unfortunately switched to “OXR1” in at least 79 papers in PubMed.
Unfortunately in this instance, it did not seem helpful to change the approved OXR1 symbol to something different, but to try to minimize future confusion, we wrote to all authors who have published on HCRTR1 with the “OXR1” alias, reiterating the importance of using approved gene nomenclature in their papers.
«««< HEAD For further examples, please read the full paper. ======= For more examples see our paper.
57501fa3a7dbc1323d7b3996e0a6a0e744f2d7d0
The consequences of using unapproved gene symbols can include the following:
-
Difficulty in finding relevant papers in the literature
-
Lack of certainty in regard to which gene a paper is about
-
Use of the wrong protein in experiments
-
Papers being incorrectly associated with the wrong gene in databases
-
Antibody companies incorrectly labelling their products
-
Clinical harm to patients
We suggest that in order to avoid confusion researchers can take the following steps:
-
Use HGNC approved nomenclature in your manuscripts - our multi-symbol checker tool can help with this.
-
Reference our stable HGNC IDs to remove ambiguity when referring to genes that have been previously published with confusing alias symbols.
-
Find the HGNC gene report for your gene of interest at genenames to view its known aliases and to see if we have added a “curator note” to highlight issues such as potential symbol and alias clashes. Alias symbols and names are searchable too!
We are keen for our blog readers to contact us if you are aware of further examples of confusion caused by clashes between approved gene symbols and aliases, beyond those discussed in our paper. For genes in non-human vertebrates, we advise that symbols approved by the relevant species-specific nomenclature committees (e.g., MGI for mouse) or the Vertebrate Gene Nomenclature Committee (VGNC) are used.
