Taking the wider perspective - the value of gene groups
Guest Post ·

Most biological databases are organized around individual pages for each gene (or protein or transcript etc). Addition of information to those databases is usually guided by the same principle - e.g. biocurators associate new data with specific genes as reported in individual research papers. While this is obviously logical and useful, a purely gene-by-gene approach also has its shortcomings: it can produce inconsistencies in the functional annotation and nomenclature of related genes, and it doesn’t allow for user-friendly presentation of lists of gene families or other functional groupings. This is where taking a wider perspective really helps, and it is the basis of the ‘gene group’ resources available at FlyBase and the HGNC.
Gene groups have been central to the HGNC since its inception, ensuring that related sets of human genes are named logically and systematically while also providing researchers with easy-to-use gene lists. Inspired by this approach, FlyBase started curating Drosophila gene groups in 2012 and, after finessing our strategy and accumulating a decent bolus, gene group report pages debuted on our website in 2015 (Attrill et al. 2016). We began with small, established groups such as Wnts, actins, and tubulins. We then expanded the resource to include other, often larger, functional groupings that were described in papers/reviews or requested by users, such as ion channels and transcription factors. More recently we have adapted our original gene group approach to undertake a systematic review of enzymes (Garapati et al. 2019) and accommodate signaling pathways (Larkin et al., 2021). As of December 2021, there are over 1,600 gene group reports in FlyBase, comprising 8,267 (46%) of all sequence-localized Drosophila genes.
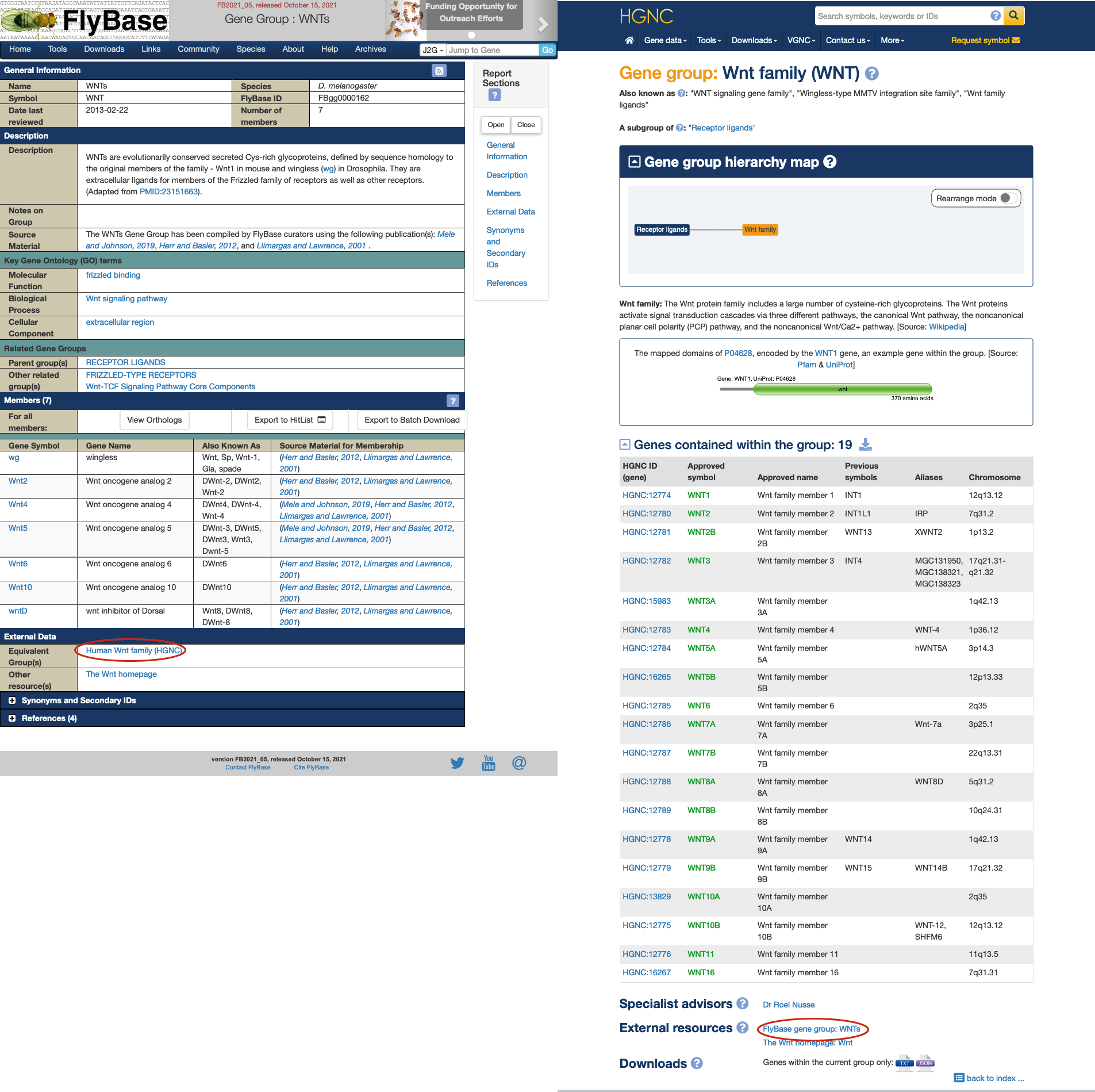
The provision of ready-made, manually verified gene groups within biological databases is useful for several reasons. First, it eliminates/minimizes the work a user would otherwise have to do to assemble the list themselves, either from multiple source publications or by navigating several different database tools. Second, the lists are coupled to the wealth of other information within the database, which also means that all the information (including gene symbols) is kept up-to-date. Third, it facilitates comparison of equivalent gene sets between species - for example, there are currently >400 reciprocal links between FlyBase and HGNC gene group reports. FlyBase gene group reports are demonstrably popular with our users, receiving the highest number of page views of any FlyBase data class (when normalized for page number) and being directly cited in over 20 research papers.
Production of web reports is only one benefit of curating gene groups. Annotation using the Gene Ontology (GO) is the primary method used by FlyBase (and most other biological databases) to describe the functions of gene products, and assembling a gene group enables the consistency and coverage of GO terms applied to the group members to be reviewed - missing GO annotations are added, any incorrect annotations are edited/removed, and improvements to pipelines generating computational GO annotations are made. The end result is that scientists searching FlyBase or other third party databases using the GO receive more accurate and complete results, in agreement with the data presented in FlyBase gene group reports.
Gene group curation also allows us to review and rationalize the nomenclature of related Drosophila genes. As alluded to above, fly genes have traditionally been named on a paper-by-paper basis, often without reference to existing naming conventions for related genes in Drosophila or other species. This can result in a frustratingly haphazard nomenclature within a gene set. A good example of this are the fly genes encoding the deeply conserved subunits of RNA polymerases (RNAPs): until recently, these were variously named in FlyBase according to their molecular weight, their yeast orthologs, their human orthologs or a protein-protein interaction, and several hadn’t been identified at all. In curating the RNAP gene group, we worked with expert researchers in the field to agree on and apply the systematic nomenclature to the fly RNAP subunits that was already in widespread use for human and vertebrate orthologs (Marygold et al. 2020). Implementation of this standardized nomenclature makes it much clearer that these genes are related as well providing information about their function and orthology relationships.
We are always keen to work with other experts to produce/improve gene group reports and in so doing also enhance GO annotation and/or review gene nomenclature - just let us know using the Contact FlyBase form.
My thanks to the FlyBase gene group curators past and present: Helen Attrill, Phani Garapati, Alix Rey and Giulia Antonazzo.
