Gene Group Jamboree 2022
HGNC ·
Photo: Springtime daffodils
The HGNC curators recently held a two day “gene group jamboree” where we reviewed our existing gene group resource and thought about how we can make it even more useful to researchers in future. There are currently 1642 gene groups that have all been manually curated.
One major change we would like to make to our gene groups is being able to classify them into types. Gene groups could be labelled with more than one type. These may include:
- shared homology
- shared structure
- shared biological activity
- complex subunits
- locus type
- community defined
- nomenclature grouping
- historical grouping
We also considered the option of using “family” and “shared domain” as labels. However, the term “family” is used in a variety of ways in the literature: some researchers may use it for a set of genes encoding proteins that share a domain, although they may not all be paralogs, or may choose to refer to a functional group as a family. The term “shared homology” infers shared evolutionary history - the genes referred to may be paralogs, or may encode proteins that share one or more domains.
We also plan to define the relationships that exist between groups in our hierarchy (such as is_a, part_of ). Another new feature may involve the use of GO terms to show which genes encode active proteins in particular groups, e.g. enzymes.
Here are a few gene group examples with discussions about what “types” we may assign them. We envisage our gene group labels/tags to have a similar appearance to our existing “Placeholder symbol” and “Stable symbol” luggage tags.
* DASH family
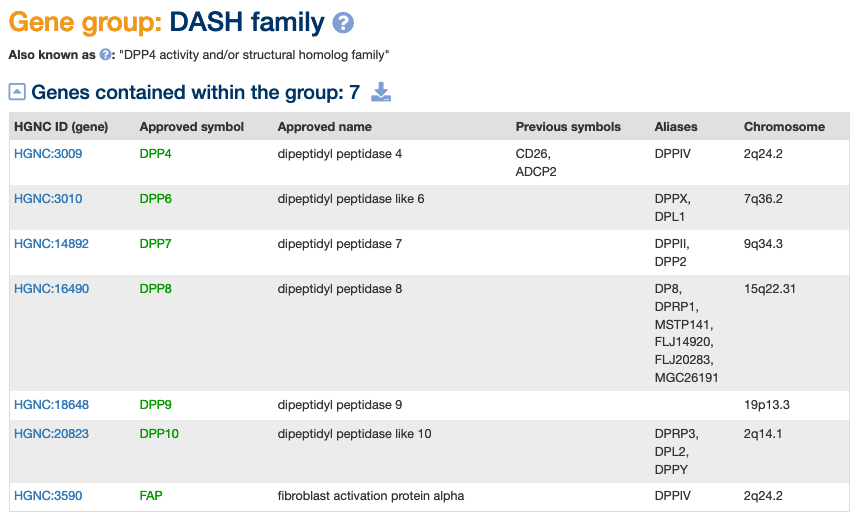
The DASH family, also known as the “DPP4 activity and/or structural homolog family” consists of genes encoding proteins with dipeptidyl peptidase activity and/or structural similarity to other DASH proteins (PMID:26671446). DPP7 (previously DPP2) encodes a protein that displays DPP4-like activity and is included in the literature as a DASH family member, but it does not show sequence similarity to the other DASH proteins and is also structurally unrelated.
PRCP (prolylcarboxypeptidase) and PRSS16 (serine protease 16) are paralogs of DPP7. PRCP and PREP (prolyl endopeptidase) are included as part of the “more broadly defined DPP family” in (PMID:26300881).
DPP6 (dipeptidyl peptidase like 6) and DPP10 (dipeptidyl peptidase like 10) encode proteins that lack enzymatic DPP4‐like activity, hence the inclusion of the word “like” in their gene names.
Although we could separate some of the genes in the DASH group into subgroups, this could create issues about where to put genes such as PRSS16, APEH and PREPL which are paralogs of some of the DASH protein encoding genes but are not included as part of this grouping in the literature. Therefore, we may choose to:
-
Keep all the genes referred to in the literature as the “DASH family” together in a single group and label as “community defined”.
-
Create additional separate groupings based on classifications in the peptidase database MEROPS: “Peptidase subfamily S9A” for DPP4 and its paralogs, “Peptidase subfamily S28” for PRCP, PRSS16 and DPP7 and “Peptidase family S9” for PREP, PREPL and APEH. These could all then be labelled as “shared homology” groups.
* Transforming growth factor beta family (TGFB)
This group contains three close paralogs, TGFB1 (transforming growth factor beta 1), TGFB2 (transforming growth factor beta 2) and TGFB3 (transforming growth factor beta 3). This is a subgroup of the transforming growth factor superfamily, which contains three subfamilies and fifteen other genes.
We could label this gene group as “shared homology”.
* LEM domain containing (LEMD)
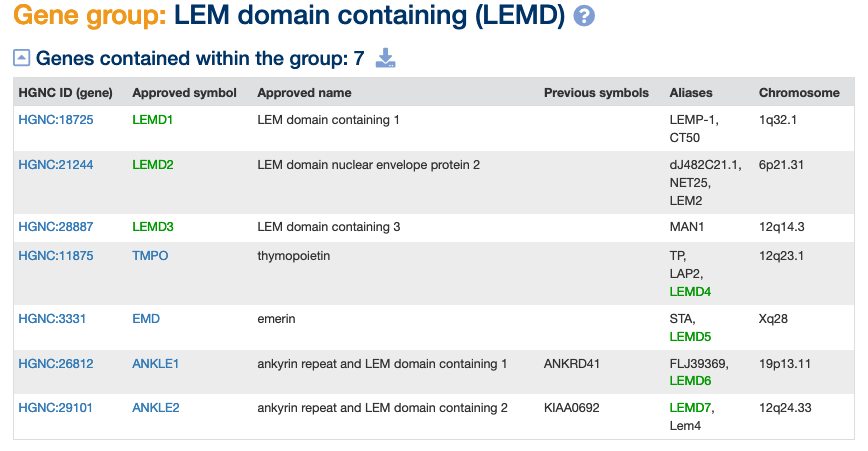
This group of seven genes has three genes named using the LEMD# root. The LEM domain name is derived from “LAP2-emerin-MAN1 domain”. “LAP2” is an alias symbol for TMPO (thymopoietin), “emerin” is encoded by EMD and “MAN1” is an alias for LEMD3 (LEM domain containing 3). Proteins containing this domain can bind lamins and tether repressive chromatin at the nuclear periphery. They interact with the protein encoded by BANF1 (BAF nuclear assembly factor 1).
We could label this gene group as “shared homology”.
These genes can be further divided into three groups based on membrane topology and other features: EMD, TMPO and LEMD1 as group 1, LEMD2 and LEMD3 as group 2 and ANKLE1 (ankyrin repeat and LEM domain containing 1) and ANKLE2 (ankyrin repeat and LEM domain containing 2) as group 3, as published in (PMID:25863918). Alternatively we could subdivide the group based on paralogy which would place LEMD1 and TMPO in a subgroup and LEMD2, LEMD3 and ANKLE1 in another subgroup. Neither EMD nor ANKLE2 have paralogs so would be in subgroups with one member each. In cases like this it can be difficult to judge which groupings are most useful and whether there is value in presenting multiple options.
* SCF complex
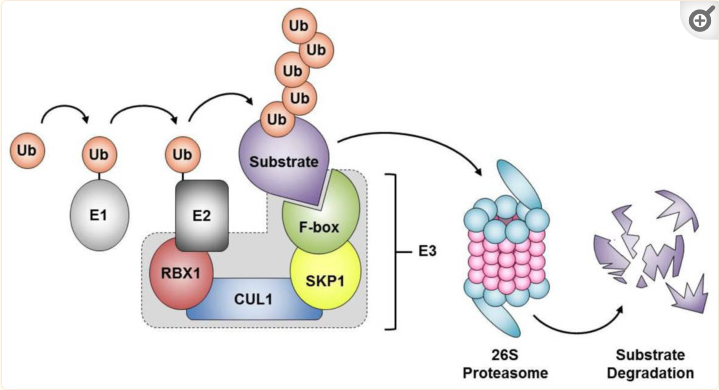
Figure from Thomson et al, Int. J. Mol. Sci. 2021, 22(16), 8544; https://doi.org/10.3390/ijms22168544. (PMID:34445249) The SCF complex orchestrates proteolytic degradation by the 26S proteasome. Schematic of the SCF complex consisting of four components, including invariant core members (SKP1, RBX1 and CUL1) and one of the 69 variable F-box proteins. The F-box proteins facilitate the transfer of ubiquitin from the E2-conjugating enzymes onto protein substrates, where poly-ubiquitination often denotes those targeted for proteolytic degradation via the 26S proteasome. E1, E1 ubiquitin-activating enzyme; E2, E2 ubiquitin-conjugating enzyme; E3, E3 ubiquitin-protein ligase; Ub, Ubiquitin.
This is a multi-protein E3 ubiquitin ligase complex catalyzing the ubiquitination of proteins destined for proteasomal degradation. The SCF complex can be divided into genes encoding proteins with F-boxes and those without.
The SCF complex core subunits group contains three genes: CUL1, RBX1 and SKP1. This group could be tagged as “complex subunits”.
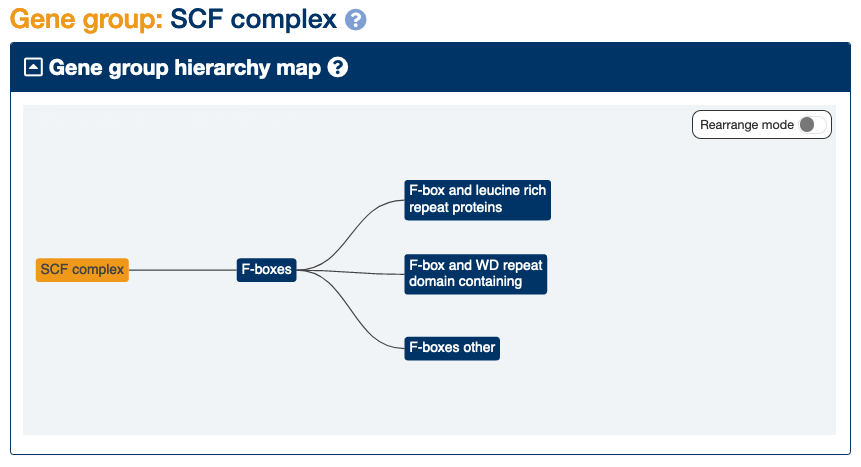
The F-boxes, the alternative SCF complex subunits, divide into three further subgroups: genes encoding proteins with an F-box and leucine rich repeats (21 genes), those with an F-box and WD repeats (11 genes) and others that just contain F-boxes (39 genes). All these subgroups could be tagged as “shared homology” and “complex subunits”.
7BS protein arginine methyltranferases
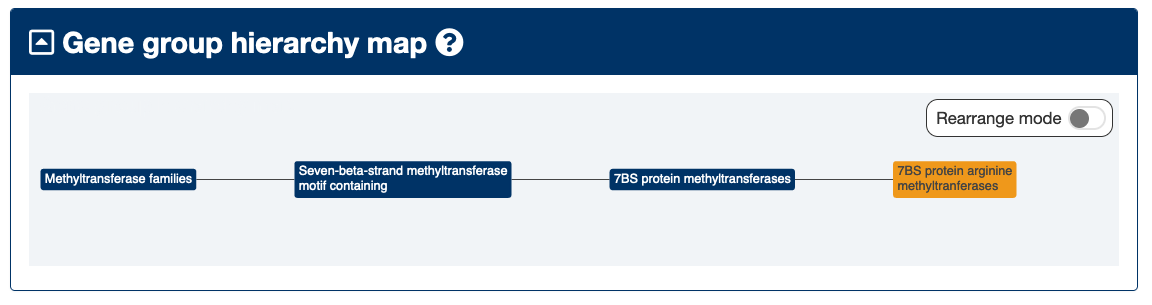
This group contains genes that encode proteins with a shared 7BS motif. All of the PRMT# root genes including PRMT1 (protein arginine methyltransferase 1) and CARM1 (coactivator associated arginine methyltransferase 1)(aliased as PRMT4) are listed as a set of paralogs in Ensembl. NDUFAF7 (NADH:ubiquinone oxidoreductase complex assembly factor 7) is not listed as having any paralogs in Ensembl.
All genes in this group encode proteins with arginine methyltransferase activity. This group could be tagged as “shared function”. If we wanted to also label this group using a shared homology tag we would need a system to show that NDUFAF7 is an exception and not a paralog of the other gene group members. This could be something we may discuss further at a future gene group jamboree.
We would love some feedback from users about what developments you might like to see for our gene groups in future. Our curators work hard to keep these up to date with the literature as far as possible, but always appreciate input from experts if you notice any gene groups are incomplete or currently missing from in our resource. Please contact us at hgnc@genenames.org.
