Gene Curation Coalition Flagship Paper
HGNC ·
We are excited to announce the paper describing the Gene Curation Coalition (GenCC)’s global effort to harmonise gene–disease evidence resources is now published in Genetics in Medicine (available via this link until June 23rd 2022).
Gencc.org gathers gene-disease associations from many sources
Figuring out which genes to test when diagnosing a disease can be challenging. The first problem is that there are many different clinical resources curating links between genes and diseases. Each has their own database and their own focus, but some genes will appear on several sites - checking each individually is time consuming. Diagnostic laboratories may also have relevant information that is not publicly available.
A primary goal of the GenCC was to gather all of this data onto a single website and encourage those who may not have previously shared data to do so. This is now available at gencc.org which has contributions from online resources (e.g. ClinGen, DECIPHER, Genomics England PanelApp, OMIM, Orphanet, PanelApp Australia, TGMI’s G2P), as well as diagnostic laboratories that have committed to sharing their internal curated gene-level knowledge (e.g. Ambry, Illumina, Invitae, Myriad Women’s Health, Mass General Brigham Laboratory for Molecular Medicine). In May 2022, the database contained 16,130 gene-disease associations for 4,629 different genes.
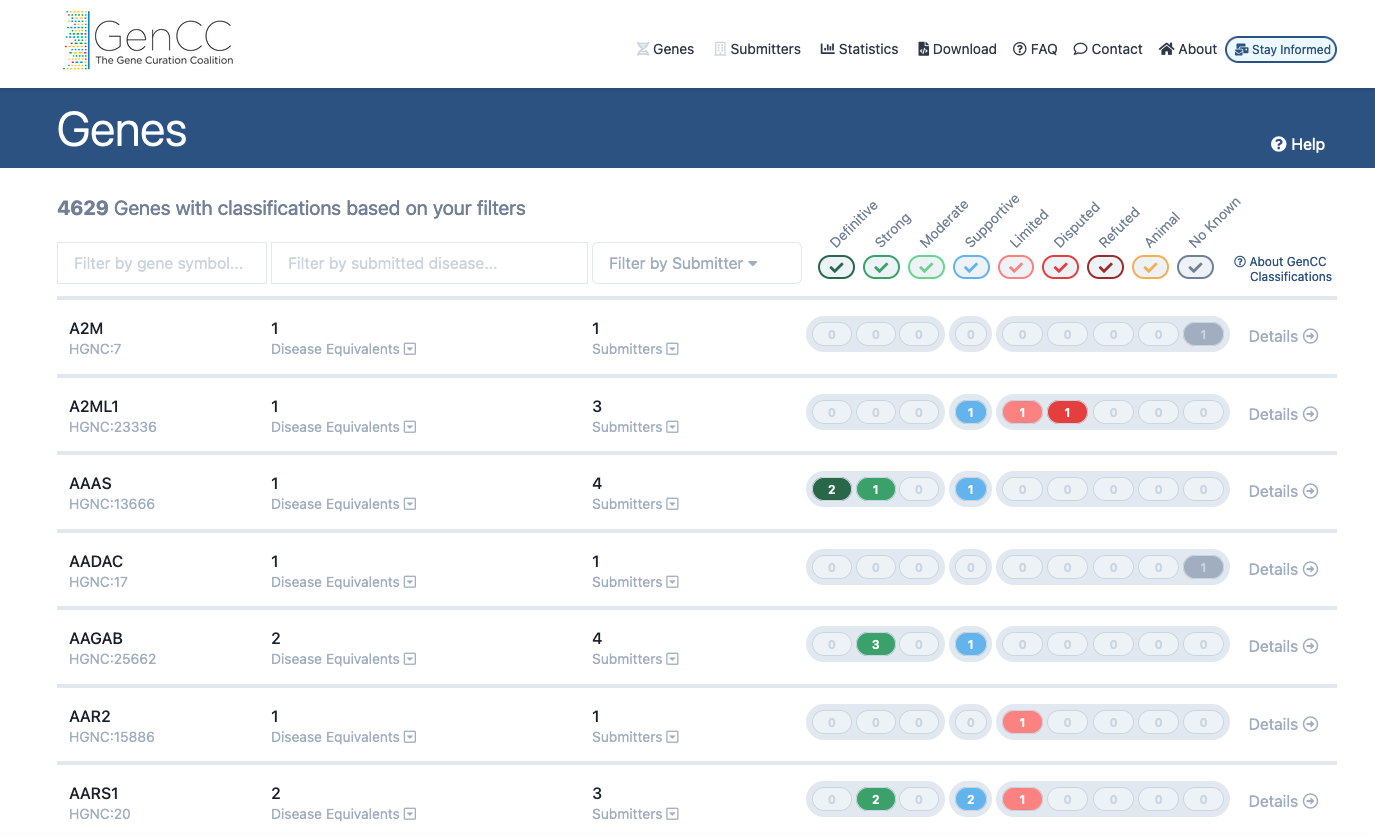
Screenshot to show a subset of gene data including GenCC classifications as displayed on the GenCC website
Harmonized terms to describe evidence for gene-disease associations
A second problem when assessing genes for testing has been that different resources used different ways to describe the evidence supporting the disease association. One resource may say an association is ‘possible’, another that the evidence is ‘limited’ while a third shows a coloured-coded ‘amber’ traffic light. Are they saying the same thing? Different terms can be confusing when you are trying to decide if a gene is truly causative of a disorder or not. GenCC has addressed this by getting everyone involved to agree on a consistent set of terms to use. These range in strength from ‘definitive’ through ‘strong’, ‘moderate’, ‘supportive’, ‘limited’, ‘disputed’, ‘animal model only’, ‘refuted’ and ‘no known evidence’. Each has a definition so they can be applied consistently. The paper includes a description of the Delphi survey that was used to reach agreement on evidence terms. Some clinical resources have switched to using these evidence classifications on their own website and others are currently mapping to the agreed terms when they submit data to GenCC.
Resolving conflicting opinions
What do you do when one resource is confident of a gene-disease association and another isn’t? Minor differences in opinion between groups are to be expected - maybe one hasn’t seen a recent paper that would bump the evidence up from ‘strong’ to ‘definitive’ - however serious conflicts such as ‘strong’ versus ‘refuted’ need to be addressed. GenCC has regular meetings where the submitting groups can discuss and resolve such conflicts. Users can also view the supporting evidence used to make each association. There may also be differences in the choice of disease terms used by different groups - some go for gene specific terms whereas others favour more generic terms e.g. Orphanet link ABCB6 to ‘microphthalmia, isolated, with coloboma’ but TGMI G2P and PanelAPP Australia link the same gene to the more specific term ‘microphthalmia, isolated, with coloboma 7’ which is defined as due to a mutation in ABCB6. Consequently, the next step is to work on harmonising the choice of Mondo disease ontology terms used by the different groups.
Stable symbols for disease genes
When disease and patients are involved, it is vital that researchers, clinicians and curators of clinical resources are in no doubt which gene is being discussed. The GenCC displays HGNC approved gene symbols and HGNC IDs to make this clear. HGNC is a member of GenCC - although we don’t make gene-disease associations we have committed to reviewing the nomenclature of each gene in the GenCC database. We make sure the language used is appropriate for clinical settings and not offensive or pejorative in any way. We also look for anything that is incorrect or potentially misleading in the gene name or symbol. Where necessary we update the gene name or, in exceptional cases, change the gene symbol. Once we are sure a symbol is unlikely to change again we mark it as stable. We are using the GenCC evidence categories to prioritise our nomenclature review - so far we have stabilised symbols for 70% of the 1,621 genes that have at least one definitive disease association in the GenCC database and 54% of all gene symbols in GenCC. Our stable symbols are marked with ‘stable symbol’ luggage tags at the top of our gene report pages. Further down the gene report we link to Clinical Resources including GenCC.
Breaking news! HGNC/VGNC user feedback survey 2022 is now live
Please fill in our survey and let us know what you think of the genenames.org resources. This is particularly important for us at this point in our funding cycle and we appreciate you taking the time to provide us with some feedback.
