CD molecules
HGNC ·
CD molecules as cell surface markers
If you read immunology papers, you will have no doubt come across many mentions of “CD (cluster of differentiation) molecules”, also known as CD antigens. CD molecules can be used as cell surface markers to enable researchers to differentiate between cell types. CD molecules can be recognized by particular monoclonal antibodies (mAbs) and the CD designation refers to both the group of mAbs that recognize a cell surface molecule, and to the molecule itself (PMID:26546687). The addition of a + or - sign in superscript can be used to refer to a particular cell population that displays (+) or lacks (-) the CD molecule in question. Cell sorting methods such as flow cytometry can be used to separate cells expressing particular markers.
Although CD molecules were initially used as surface markers to differentiate between different populations of leukocytes (including T cell and B cell lymphocytes, basophils, neutrophils and eosinophils), the system has now expanded to include markers found on some other cell types as well, including erythrocytes, platelets, endothelial cells and epithelial cells.
Some examples of cell populations that can be identified include:
-
CD4+ T helper cells - MHC class II molecules present peptides to CD4+ T cells. The protein encoded by CD4 (CD4 molecule) is a co-receptor for the T-cell receptor complex (TCR). T helper cells release cytokines which regulate the activity of other immune cells.
-
CD8+ T cytotoxic cells - MHC class I molecules present peptides to CD8+ T cells. CD8 is a dimeric co-receptor for the T-cell receptor TCR. This co-receptor can be a heterodimer, composed of CD8A and CD8B encoded protein chains, or a homodimer of alpha chains only.
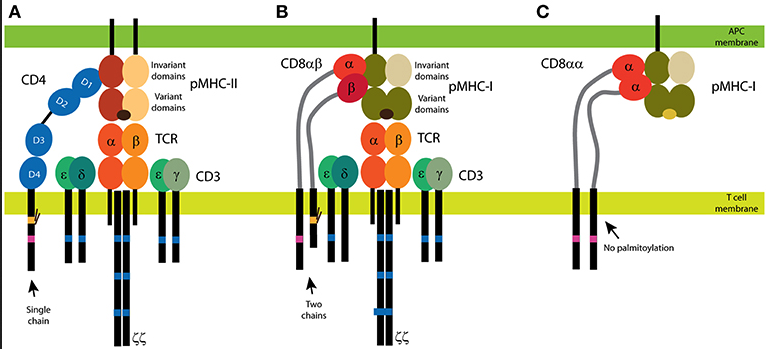
Figure 1. from Glatzová and Cebecauer (PMID:31001252).
CD4 and CD8 coreceptors. (A) The CD4 glycoprotein is composed of a single chain. Its functional motifs, such as the Lck-binding site (in magenta) and the palmitoylation site (in yellow), are in the sole intracellular domain. The extracellular part of CD4 is composed of four Ig-like domains, and the MHC binding site is in the N-terminal D1 domain. Short linker connects CD4 extracellular domains with the transmembrane domain. (B,C) Two forms of CD8 exist: the αβ heterodimer (B) and the αα homodimer (C). The α subunit of CD8 contains the Lck-binding site, and the β subunit contains the palmitoylation site. A single Ig-like domain and a long stalk region (in light gray) form the extracellular parts of the CD8 subunits. Binding of CD4 (A) and CD8αβ (B) to MHC is illustrated with the antigenic receptor because these coreceptors support receptor function in T cells. The TCR/CD3 complex is composed of at least eight subunits. CD3 subunits γ, δ, and ε contain one immunoreceptor tyrosine-based activation motif (ITAM; in dark blue) and three ITAMs are in each ζ subunit. Cognate peptides are depicted in dark brown, self-antigens in light brown.
- CD45+ - This is used as a marker of all hematopoietic cells except for mature erythrocytes and platelets. The HGNC has this gene approved as PTPRC (protein tyrosine phosphatase receptor type C) with CD45 listed as an alias.
- CD47+ - This plays a role as a regulator of integrin activation, and in erythrocytes links the protein encoded by SLC4A1 (solute carrier family 4 member 1 (Diego blood group)) and also referred to as “erythrocyte membrane protein band 3” and the Rhesus complex, which consists of the Rhesus antigens encoded by RHD (Rh blood group D antigen) and RHCE (Rh blood group CcEe antigens) along with many accessory proteins, which helps to maintain the integrity of erythrocyte membranes. It also functions as a marker of self on erythrocytes through binding the protein encoded by SIRPA (signal regulatory protein alpha). This has an inhibitory effect, making SIRPA expressing dendritic cells and macrophages less likely to phagocytose autoimmune sensitized cells with CD47 on the surface (PMID:15359629).
Some antibodies that identify CD molecules can be used diagnostically. The abnormal expression of CD markers in the bone marrow and blood of patients is one of the first diagnostic features of leukemia (PMID:32782727). Around 85-90% of patients with acute myeloid leukemia (AML) and 95-100% of patients with acute promyelocytic leukemia (APL) are said to be “CD33-positive” (PMID:34203180) However, this does not mean that CD33 is expressed on every cancerous cell - if it is expressed on 25% or more leukemic blasts then the case is classfied as being CD33-positive. CD33 (CD33 molecule), also published using the alias SIGLEC3 encodes a lectin protein that binds sialic acid and acts as a transmembrane receptor on hematopoietic cells (PMID: 7718872). The CD33 encoded protein is an inhibitory receptor that recruits the proteins encoded by PTPN6 (protein tyrosine phosphatase non-receptor type 6, alias SHP-1) and PTPN11 (protein tyrosine phosphatase non-receptor type 11, alias SHP2) upon phosphorylation, leading to a downregulation in the levels of inflammatory cytokines released by immune cells.
Single nucleotide polymorphisms (SNPs) in genes encoding CD molecules can also be studied and used as a diagnostic tool. For example, a particular variant of the CD38 gene can be used as a marker for chronic lymphoblastic leukemia (CLL). Patients carrying the rs6449182 GG genotype have a higher expression level of CD38 protein and progress to higher clinical stages relative to patients lacking this genetic change (PMID:32782727).
Expression of TNFRSF8 (TNF receptor superfamily member 8) alias CD30, and its ligand TNFSF8 (TNF superfamily member 8) alias CD30L is upregulated in patients with several hematological cancers and other inflammatory conditions including lupus erythematosus, rheumatoid arthritis, asthma and atopic dermatitis (PMID:19760074).
Monoclonal antibodies generated against certain CD markers can be used therapeutically in cancer treatment. These treatments include:
-
rituximab for CLL (targets the protein encoded by MS4A1 (membrane spanning 4-domains A1) (alias CD20))
-
daratumumab for multiple myeloma (targets the protein encoded by CD38 (CD38 molecule))
-
trastuzumab (Herceptin) for breast and stomach cancers (targets the protein encoded by ERBB2 (erb-b2 receptor tyrosine kinase 2), alias HER2 and CD340)
-
gemtuzumab ozogamicin (brand name Mylotarg)
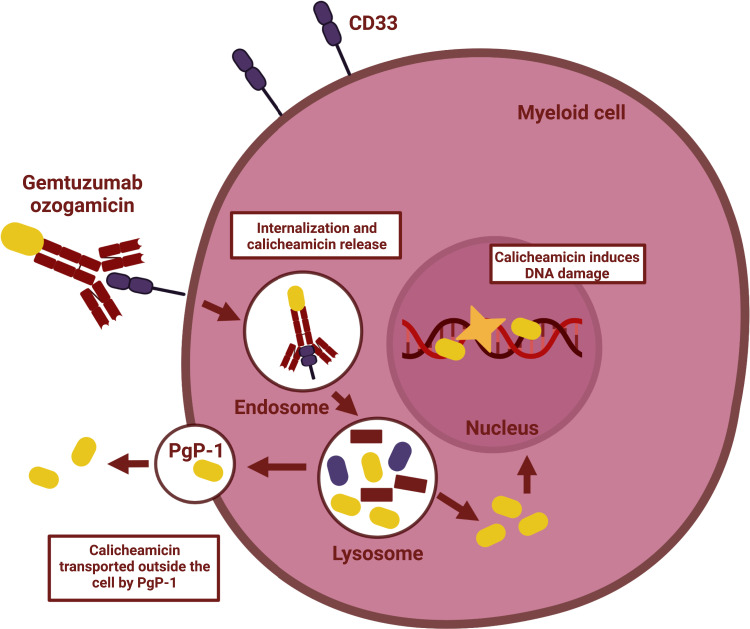
Figure 1 from Wijnen et al Onco Targets Ther. 2023; 16: 297–308. PMID:37153641
Gemtuzumab ozogamicin is a CD33 protein targeting antibody-drug conjugate used to treat acute myloid leukemia (AML). Its approval for use was withdrawn in 2010 because of safety concerns, with some patients taking it shown to be at an increased risk of experiencing a condition known as veno-occlusive disease (VOD), a serious condition where blood vessels around and in the liver become blocked. There were also other serious potential side effects of this treatment recorded including hemorrhages and acute respiratory distress syndrome (PMID:31308990). Further trials had to be done before the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) reversed this decision in 2017 to allow this treatment to be used in some patients. These later studies concluded that the benefits of gemtuzumab ozogamicin outweighed the risks for elderly patients who could not tolerate other more intensive chemotherapy. It is less likely to be used in patients who are likely to get an allogenic stem cell bone transplant because of the risk of VOD.
CD molecule Nomenclature
The organisation Human Cell Differentiation Molecules (HCDM) runs workshops to name and characterise CD molecules. The 1st International Workshop and Conference on Human Leukocyte Differentiation Antigens (HLDA) was held in Paris in 1982 to propose and establish the nomenclature. A CD cluster is only assigned a CD number when at least two specific mAbs have been shown to bind the molecule in question. The complete current list of CD molecules assigned through HLDA workshops can be seen here.
There are currently 394 genes in the HGNC CD molecules gene group. Of these, 76 genes are currently approved using CD symbols, but 318 are approved with other symbols with CD aliases. The CD molecules do not represent one large gene family - they did not evolve on cell surfaces for the convenience of researchers! Therefore it makes sense that they are members of many different gene families and their encoded proteins have a variety of molecular functions.
We are now aiming to stabilise gene symbols when possible, and 21 genes have currently had their nomenclature stabilised with approved CD symbols.
As they have the shared feature of being cell surface expressed, genes included in the CD molecule group commonly have roles in
-
cell signalling, acting as receptors or ligands e.g. KIR2DL1 (killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 1) (alias CD158A), PLAUR (plasminogen activator, urokinase receptor) (alias CD87), CXCR1 (C-X-C motif chemokine receptor 1, alias CD181) and CD40LG (CD40 ligand).
-
cell adhesion (e.g. ALCAM, alias CD166 and ICAM (intercellular adhesion molecule 4 (Landsteiner-Wiener blood group).
-
functioning as cell surface enzymes (e.g. MME (membrane metalloendopeptidase) (alias CD10) and ANPEP (alanyl aminopeptidase, membrane) (alias CD13).
Multiple factors can affect which of these genes have been named as CDs and which have alternative approved symbols, including:
-
Usage in the literature (We are now looking to stabilise well used gene symbols when possible).
-
What is known about the molecular function of the protein product encoded by the gene
-
Whether the gene is known to be part of an evolutionarily related family
It is quite common for the literature to favour the usage of CD symbols or alternative symbols depending on the biological field: immunologists tend to use CD numbers a lot in their papers, whereas those in other fields may choose to use alternative nomenclature. The HGNC strives to encourage the usage of approved symbols and ask that researchers use or at least mention the approved nomenclature in their publications, to minimise confusion and aid data retrievability.
We are keen to hear your thoughts on our CD molecule nomenclature. Should we add extra functional information into the names of more of these genes, or is it preferable to keep the names short and simple? For example, we have CD3D named as “CD3 delta subunit of T-cell receptor complex” whereas CD2 is currently named as “CD2 molecule”. This name could be updated to “CD2 molecule T-cell adhesion mediator” for example, (PMID:17172599) while retaining its well published CD2 symbol.
Are there any genes currently named as CD molecules that you would be keen to see renamed? We can only do this if the community working on a gene is highly supportive of a change and the gene symbol in question is not already entrenched in the literature, or already has the stable tag. However, we would be happy to hear your thoughts on any proposals you may have.
