Autumn newsletter 2019
Newsletters ·
Thanks to our Scientific Advisory Board
We would like to thank all the members of our SAB for attending our annual meeting, held here at EMBL-EBI from 7-8 November. We were pleased to welcome two new board members: Professor Anne Ferguson-Smith is the Arthur Balfour Professor of Genetics and Head of the Department of Genetics at Cambridge University. She brings her expertise in mammalian developmental genetics, which is invaluable to both the HGNC and VGNC projects. Professor Peter Robinson is the Donald A. Roux Chair in the Genomics and Computational Biology department of the Jackson Laboratory for Genomic Medicine in Bar Harbor, Maine, USA and is Principal Investigator of the Monarch Initiative, a project that aims to integrate genotype-phenotype data across species and has developed the Human Phenotype Ontology as part of this work.
We also enjoyed a pre-meeting with our external gene family collaborators: Tsviya Olender of the Weizmann Institute in Israel who works on olfactory receptor (OR) nomenclature, and David Nelson of the University of Tennessee and Jed Goldstone of the Woods Hole Oceanographic Institution in Massachusetts who both work on cytochrome P450 (CYP) nomenclature. As discussed below, we have started to add gene groups into the VGNC resource based on their work.
New VGNC website
We are delighted to announce that we have just released a new version of our VGNC website vertebrate.genenames.org! The redesign brings the VGNC website in-line with the current HGNC site, both in terms of the look and feel of the site and the underlying web code and technologies used.
New features of the homepage include:
- A tagline ‘The resource for approved gene nomenclature in selected vertebrates’ where the term ‘selected vertebrates’ now links through to a full list of species with VGNC-approved nomenclature. This table also lets the user know whether we are naming the ‘full species gene set’, or providing symbols and names for ‘specific gene families only’ in a particular species, and gives a full count of gene symbols for each species.
- Information on when the site was last updated underneath the main search box.
- A new footer which features a full site map. This footer is displayed on every page of the new site.
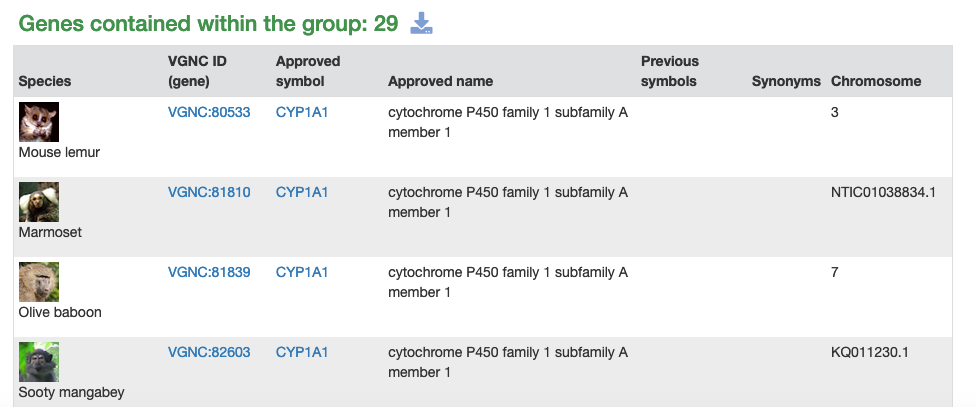
As mentioned above, we have now have a gene groups resource on vertebrate.genenames.org. This has a consistent look with the human gene groups resource on genenames.org with the obvious difference being that the VGNC gene groups contain genes from different species. As a result of this, the first column of each VGNC gene group table contains the name of the species and an image of a representative animal.
e.g.
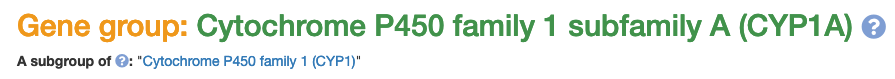
 (additional table rows not shown)
(additional table rows not shown)
To date we have gene groups for part of the cytochrome P450 (CYP) and olfactory receptor (OR) gene superfamilies.
The CYP data was provided by David Nelson and is for the full CYP superfamily gene set for 17 primate species, not including chimp or macaque. The primate superfamily contains 930 primate CYP genes sorted into more than 45 subgroups. The gene groups are sorted into a hierarchical structure that users can browse through, starting with the top-level Cytochrome P450s (CYP) page. An example subgroup is the CYP1A gene group page. We will work on adding genes for the other species in VGNC to these gene groups in the future.
The olfactory receptors superfamily currently contains curated dog genes for four major olfactory receptor subgroups: OR10, OR11, OR12 and OR13, all of which are sorted into further subgroups. This work was done in collaboration with Tsviya Olender.
The addition of gene groups into the VGNC resource has necessitated an update of the search on vertebrate.genenames.org – the search box has ‘Gene groups’ as a drop-down choice and the search results now have a ‘filter by type’ facet that allows users to select ‘Gene’ or ‘Group’. As before, you can also filter results by gene locus type and species, and there is a new option to filter based on ‘gene entry status’, which is either ‘Approved’ or ‘Entry withdrawn’ (although we do not currently have any VGNC genes with the status ‘Entry withdrawn’!).
We hope that you enjoy using the new site and, as ever, we welcome your feedback, either by emailing us directly via our VGNC email address vgnc@genenames.org, or by using the feedback form on the vertebrate.genenames.org website.
Stable tag now on genenames.org
If your favourite gene is clinically relevant, you may see our new ‘stable tag’  next to the symbol at the top of the page on the HGNC Symbol Report (see TP53 for an example). The presence of this tag means that the gene has been reviewed manually by an HGNC curator and we think that it is extremely unlikely that we would ever change the gene symbol.
next to the symbol at the top of the page on the HGNC Symbol Report (see TP53 for an example). The presence of this tag means that the gene has been reviewed manually by an HGNC curator and we think that it is extremely unlikely that we would ever change the gene symbol.
To read more about the metrics we used to decide whether a symbol can be tagged as stable, please read our recent blog post Stable symbols - improving scientific communication long-term. So far, this process has only been applied to clinically relevant genes, and is ongoing, but we do plan to extend the process beyond disease-related genes in the future.
Upcoming retirement of old URLs and FTP files
Now that the new website has been live for over a year, we plan to retire some of the URLs and FTP files from the previous site. This will take place in three months’ time. A list of URLs that will be retired can be found on our announcements page.
The following FTP files will be removed:
- ftp://ftp.ebi.ac.uk/pub/databases/genenames/alternative_loci_set*
- ftp://ftp.ebi.ac.uk/pub/databases/genenames/reference_genome_set*
- ftp://ftp.ebi.ac.uk/pub/databases/genenames/families-old2new.csv
- ftp://ftp.ebi.ac.uk/pub/databases/genenames/genefam_list.txt.gz
- ftp://ftp.ebi.ac.uk/pub/databases/genenames/hgnc2ensembl_coords.csv.gz
- ftp://ftp.ebi.ac.uk/pub/databases/genenames/hgnc_complete_set.txt.gz
- ftp://ftp.ebi.ac.uk/pub/databases/genenames/locus_groups/*
- ftp://ftp.ebi.ac.uk/pub/databases/genenames/locus_types/*
If you haven’t changed over to the newer files yet within your pipelines, please do so soon. The newer complete set can be found at:
- ftp://ftp.ebi.ac.uk/pub/databases/genenames/new/tsv/hgnc_complete_set.txt or
- ftp://ftp.ebi.ac.uk/pub/databases/genenames/new/json/hgnc_complete_set.json
The new alternative loci, reference genome files, locus group and type files can all be found under the directories:
- ftp://ftp.ebi.ac.uk/pub/databases/genenames/new/tsv/ or
- ftp://ftp.ebi.ac.uk/pub/databases/genenames/new/json/
Changes to human histone gene nomenclature
We have recently updated the gene symbols and names of our histone gene set to be as consistent as possible with a community-driven protein nomenclature scheme which you can read about in A Unified Phylogeny-Based Nomenclature for Histone Variants.
We had to take into account how to adapt this scheme to the gene level, where several genes may encode the same protein and where the period character used in the protein nomenclature system is not allowed. We solved the problem of the period character by either not including it in the gene symbols e.g. H2AB1 encodes a H2A.B histone, or by replacing it with a hyphen where necessary e.g. H1-0 encodes the H1.0 linker histone.
We consulted with members of the histone community ahead of making the changes, and based on the feedback that we received, we included reference to ‘cluster’ in the gene names of all genes present in either of the two major human histone gene clusters on 6p22 and 1q21. We did not refer to either specific human cluster in the revised gene names because some species, such as chicken, have only one histone gene cluster and we wanted to ensure that the new gene symbols are as transferrable across species as possible. Histone genes that are not present on the human histone clusters are easily distinguished by the lack of reference to cluster in the gene name.
One of the main benefits of the new gene nomenclature is that the histone gene symbols now begin with histone type: H1#, H2A# (an exception is made for the MACROH2A genes MACROH2A1 and MACROH2A2), H2B#, H3# and H4#. This has allowed us to sort our histone gene groups by type as H1 histones, H2A histones, H2B histones, H3 histones and H4 histones.
Following this renaming of human histone genes, we have also curated and named a number of dog, cow, chimp and horse histone genes using the revised nomenclature as part of our VGNC work. We will be making VGNC gene groups for these histone families, so please look out for them in the near future on vertebrate.genenames.org!
Progress on replacing placeholder symbols
We are still busy renaming genes with placeholder symbols to provide informative nomenclature. Here are some examples of placeholders that we have renamed in the last few months, along with links to their renamed VGNC orthologs:
- C5orf30 to MACIR, macrophage immunometabolism regulator chimp, horse, cow, dog, cat, macaque
- C11orf74 to IFTAP, intraflagellar transport associated protein cow, horse, dog, chimp, macaque
- CXorf40A to EOLA1, endothelium and lymphocyte associated ASCH domain 1 (no orthologs identified via the VGNC pipeline at this time)
- CXorf40B to EOLA2, endothelium and lymphocyte associated ASCH domain 2 (no orthologs identified via the VGNC pipeline at this time)
- FAM49A to CYRIA, CYFIP related Rac1 interactor A chimp, horse, cow, dog, cat, macaque
- FAM49B to CYRIB, CYFIP related Rac1 interactor B chimp, horse, cow, dog, cat, macaque
New HGNC gene groups
We have curated several new gene groups for genenames.org within the past few months as part of our ongoing curation work. Highlights include:
- IPT domain containing
- Ectonucleotide pyrophosphatase/phosphodiesterase family
- Pentatricopeptide repeat containing
- B7 family
- Guanylate binding proteins
- Profilins
- TAFA chemokine like family
- SSX family
- SPANX family
- LGI family
- Mitochondrial iron-sulfur assembly components
Gene Symbols in the News
The alleles of TAS2R38 that you carry may explain how much you like your broccoli − individuals with two copies of the allele commonly known as ‘PAV’ are particularly sensitive to bitter food, while those with one copy are more sensitive than those who do not carry this allele at all.
Individuals with loss-of-function MRAP2 gene mutations have an increased risk of obesity, high blood sugar and high blood pressure. A study performed in vitro experiments with 6 loss-of-function variants of this gene found that the MC4R signalling pathway was disrupted, leading to hopes that drugs targeting this pathway may help individuals with the MRAP2 gene mutation in the future.
The APOE gene continues to be one of the most studied and reported on in the media. A recent study suggested that an APOE variant associated with Alzheimer disease is also associated with risk of post-operative delirium in older individuals. A separate study on a woman who carried a PSEN1 gene variant associated with early onset Alzheimer disease who did not develop neurodegenerative symptoms until much later in life found that she carried two copies of the APOE ‘Christchurch variant’, leading to suggestions that this APOE variant had a protective effect.
Meeting News
Ruth attended RNA Informatics 2019 meeting in Hinxton, UK from 9-11 September where she presented a poster on ‘How to name a long non-coding RNA’. Staying on the subject of RNA, Elspeth and Ruth attended the annual RNAcentral Consortium meeting at EMBL-EBI where they enjoyed the opportunity to talk to collaborators from the RNA, annotation and model organism world.
Susan attended A Century of Genetics, a meeting that celebrated 100 years of the Genetics Society in Edinburgh, UK from 13-15 November 2019, where she presented a poster referring to our own project’s celebratory birthday this year ‘HGNC: 40 years of standardised human gene nomenclature’.
Publications
Mudge JM, Jungreis I, Hunt T, … Seal R, Bruford E. et al. Discovery of high-confidence human protein-coding genes and exons by whole-genome PhyloCSF helps elucidate 118 GWAS loci published online ahead of print, 2019 Sep 19. Genome Res. 2019;10.1101/gr.246462.118. doi:10.1101/gr.246462.118
