A Debate About Vertebrate Gene Naming
VGNC ·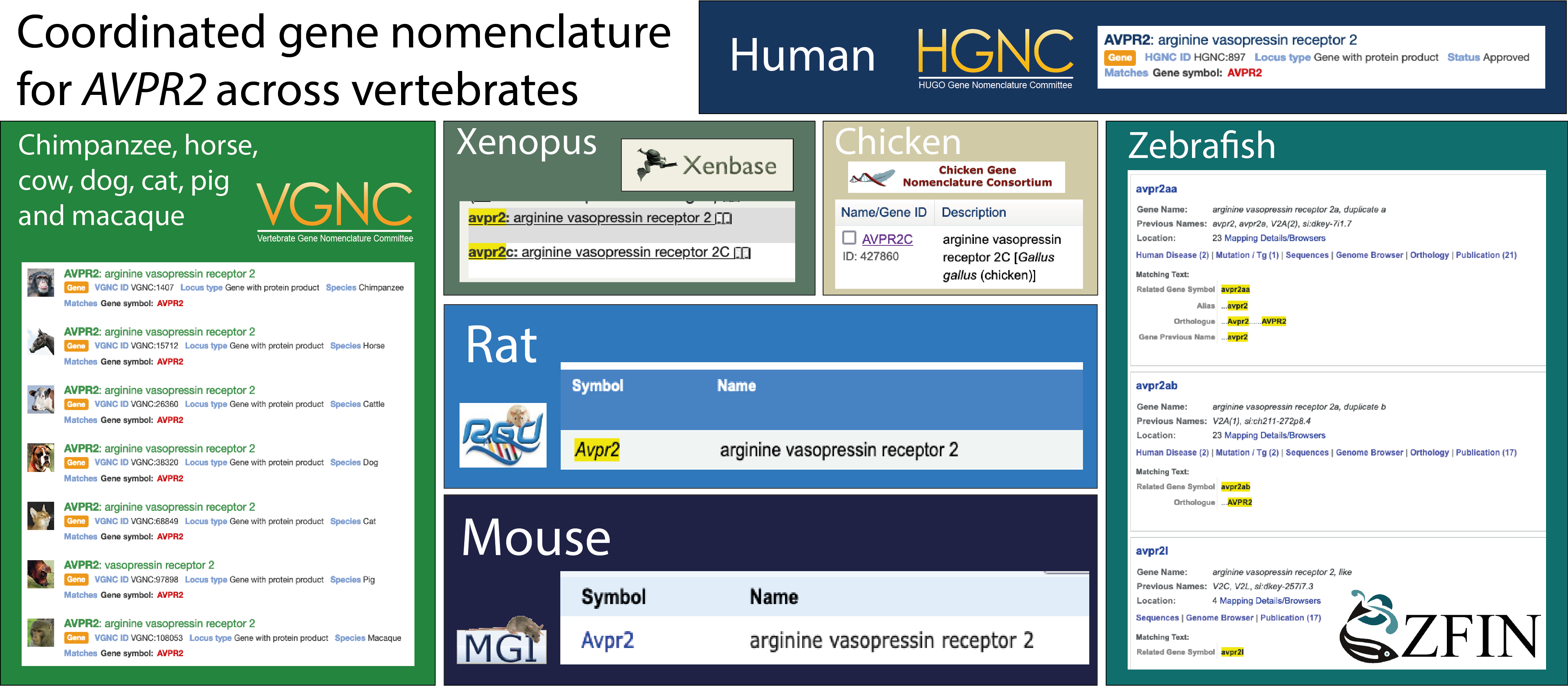
Figure 1. A summary of the gene nomenclature for the AVPR2 subfamily agreed across the vertebrate nomenclature committees.
Gene nomenclature is an essential aspect of scientific communication in biology, and has multiple functions. A gene name ideally tells us something about the gene or gene product; this might be a molecular function of the gene, its role in a biological process, or its structure. Gene symbols can be used as abbreviations to succinctly refer to specific genes, and ideally can also be used as unique search terms to help find information in publications and in databases. Consistent gene names and symbols help enable researchers and scientists to unambiguously communicate about genes and find all the associated research studies.
Another important function of gene symbols is that they can help communicate information about gene evolution. Genes that are equivalent across species are called orthologs, and we try to give orthologs the same gene nomenclature in different vertebrate species. Sometimes gene duplications have resulted in multiple copies of a gene in some species: in these cases we try to give genes the same “root symbol” with added suffixes to show that these are closely related genes. For example, CDK11 is a gene that has a single copy in most vertebrate species, but it is duplicated in humans, so the two duplicates in human are approved as CDK11A and CDK11B.
The HUGO Gene Nomenclature Committee (HGNC) was originally formed over 40 years ago to standardise gene nomenclature in humans. There are also nomenclature committees for major animal models, namely for mouse, rat, chicken, Xenopus frog, and zebrafish. All of these gene nomenclature committees have been coordinating their activities and have a long history of collaboration with the aim that genes are named consistently across vertebrate species. Gene naming across vertebrates is largely based on the human gene nomenclature, where appropriate. With an ever increasing number of genome sequences from different species available for comparative studies, gene nomenclature authorities play a crucial role in coordinating the assignment of gene names that can be propagated across species. More recently, the HGNC established a sister project to expand standardised gene naming to key vertebrate species that don’t have their own dedicated nomenclature committee: The Vertebrate Gene Nomenclature Committee (VGNC).
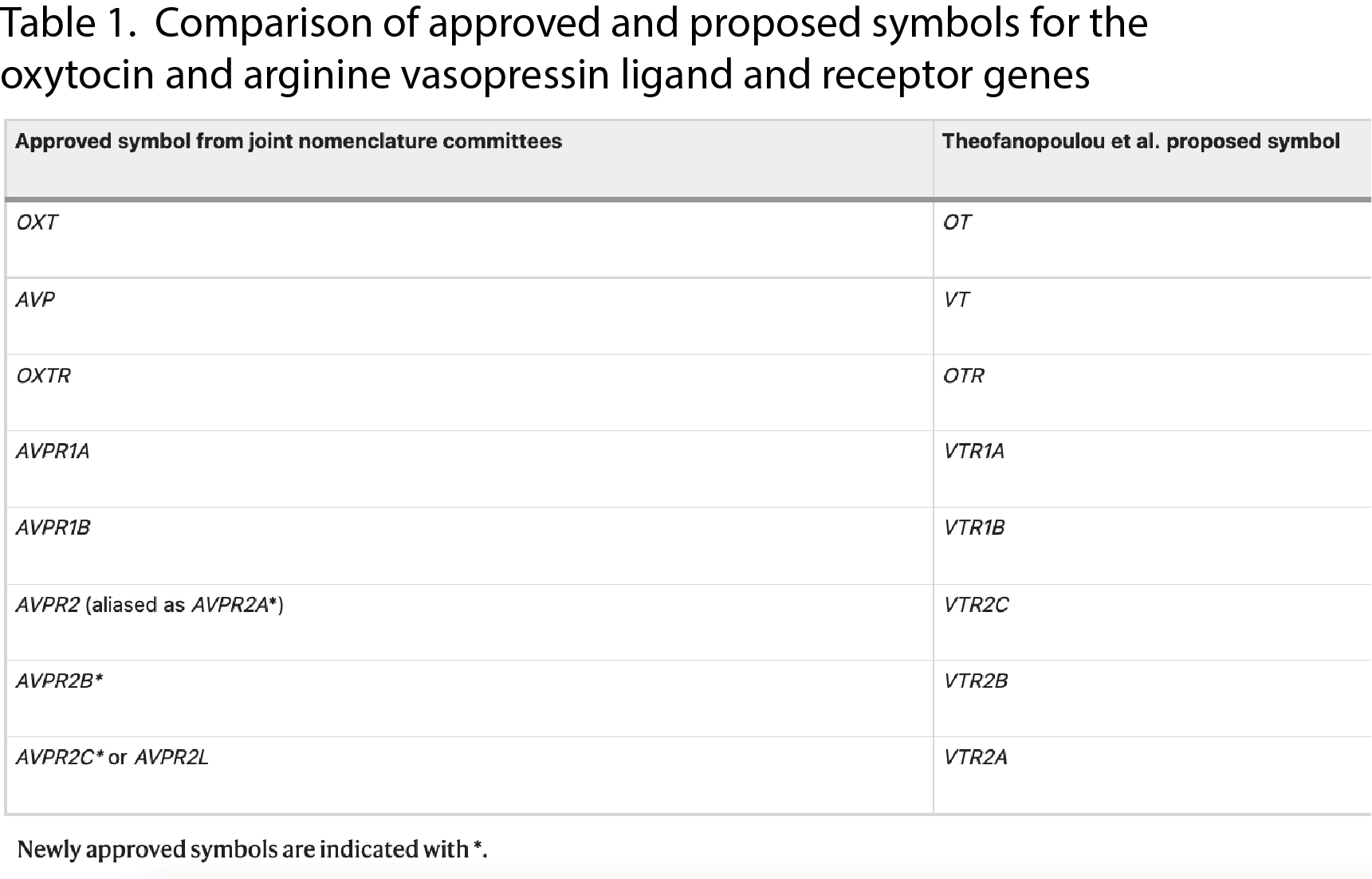
In the past few years, a project has been undertaken to sequence the genomes of all ~70,000 living vertebrate species. This project, the Vertebrate Genomes Project (VGP), is a global collaboration between researchers at several institutes and in 2021 a special issue of the journal Nature was dedicated to findings from the first wave of genomes sequenced. One of the studies in this issue (Theofanopoulou et al) was dedicated to examining the evolution of a specific set of genes: the oxytocin and arginine vasopressin genes and their receptors. The authors of that study proposed that the genes in this family be renamed based on their findings (see Table 1), and also proposed the idea of a new Universal Vertebrate gene nomenclature group, which would revise gene nomenclature using the data generated by the VGP project.
All of the existing vertebrate gene nomenclature committees, including HGNC and VGNC, collaborated on a response to this article (McCarthy et al 2023) which has just been published in Nature. In this response, we argue that dramatic changes to the nomenclature of the oxytocin and arginine vasopressin genes would be confusing for researchers because these genes are highly studied using the existing approved nomenclature. These genes are also important for human health - changes to the gene names could risk causing confusion for doctors and patients. Instead, we suggest minor updates to the current approved nomenclature of these vertebrate genes to better reflect their evolutionary history, rather than adopting a completely new nomenclature. This will help to maintain consistency and improve the tracking of information in the scientific literature. We explain that the methods and procedures used by the existing gene nomenclature committees already take into account multiple strands of information about gene family evolution, along with many other important considerations, and encourage researchers and journals to contact us before proposing changes to gene nomenclature.
An example of one of the genes discussed in these publications is shown in Figure 1. AVPR2 is a gene that encodes a receptor for arginine vasopressin (AVP). In mammals there is just one copy of AVPR2, but in other species there are varying numbers. The gene nomenclature committees have given these genes the same root symbol to indicate that they are closely related, and assigned suffixes that indicate their evolutionary history.
Ultimately, we hope that this exchange between the vertebrate gene nomenclature committees and the VGP will result in collaborations that can enable us to reflect the exciting findings from this project within the established framework of vertebrate gene naming.
You can read the full debate here.
