Spring newsletter 2021
Newsletters ·
New HGNC search application is live!
Readers of the Winter newsletter will remember being asked to test the beta version of our improved search. Thanks so much to all who did this and provided us with feedback. On April 1st we switched over to this new search on genenames.org. We have been enjoying the improved search ever since and we hope you have too! We are always happy to receive your feedback, both positive and negative…
MANE transcripts now on genenames.org
You can now find MANE (Matched Annotation from NCBI and EMBL-EBI) Select transcripts on our Symbol Reports and in our REST service. The MANE project aims to provide a set of standard transcripts for human protein-coding genes annotated by both RefSeq and Havana-Ensembl, for which there has been agreement between annotators from both teams on the entire sequence, including 5’ UTR, coding region and 3’ UTR. Please read our recent guest blog post, ‘Transcripts are the MANE attraction’ by Jane Loveland of the Havana-Ensembl team to learn more. Here is an example from our MTOR Symbol Report showing a MANE Select transcript in the ‘Nucleotide Resources’ section:
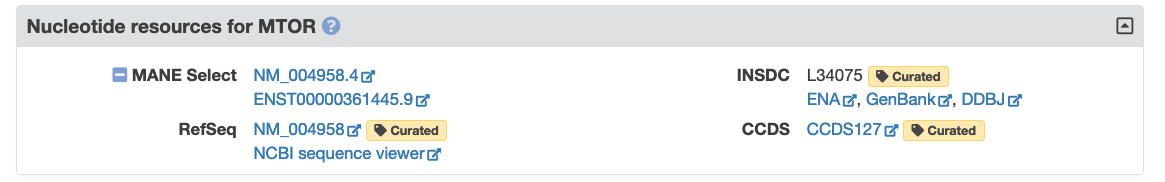
Note that MANE transcripts have both RefSeq and Ensembl IDs, and these are versioned, i.e. the MTOR gene report shows both the RefSeq ID NM_004958.4 and the Ensembl ID ENST00000361445.9. These IDs link through to transcript pages in NCBI Gene and Ensembl, respectively.
All about HCOP
We are delighted to announce that we recently published a paper in Briefings in Bioinformatics ‘Updates to HCOP: the HGNC comparison of orthology predictions tool’ describing the current version of our HCOP tool (to see the full citation for this paper, please see the ‘Publications’ section of this newsletter).
The original version of HCOP was created nearly 20 years ago and initially collated orthology calls between human and mouse from a number of orthology prediction resources, in order for the HGNC and the Mouse Genomic Nomenclature Committee to identify cases where gene nomenclature could be aligned between the two species. Two decades later, the current tool presents orthology predictions between human and the following 19 different species: chimp, rhesus macaque, mouse, rat, dog, cat, horse, cow, pig, opossum, platypus, chicken, anole lizard, xenopus, zebrafish, Caenorhabditis elegans, fruit fly, Saccharomyces cerevisiae and Schizosaccharomyces pombe using 14 separate orthology prediction resources (please see Table 1 in the new paper to view the full resource list). As many of our readers will know, we still use HCOP to align nomenclature; we now use the tool to auto-approve appropriate gene nomenclature for each VGNC full species gene set (chimp, rhesus macaque, dog, cat, cow, horse and pig). We have a software pipeline that searches the HCOP data to identify high confidence ortholog sets between human and each VGNC species, as predicted by Ensembl, NCBI Gene, OMA and PANTHER. These VGNC genes are then auto-assigned the same gene symbol as their human ortholog, provided the symbols pass rules devised by curators to ensure that the human nomenclature is suitable for transfer across species. Genes not identified in this pipeline are not given auto-approved symbols and need to be assigned nomenclature manually by a curator - a huge task which we are still working towards for each core VGNC species!
In addition to orthology calls, HCOP displays approved nomenclature from HGNC, VGNC, MGNC (mouse gene nomenclature from MGI), Rat Genome Database (RGD), Chicken Gene Nomenclature Consortium (CGNC), Xenbase, ZFIN, WormBase, Saccharomyces Genome Database (SGD) and PomBase.
One of the biggest strengths of HCOP is that it is updated daily, so the data is always as current as possible. For example, it includes the latest data from Ensembl, OMA and PANTHER, which have all released new ortholog sets in the last few weeks. Please read the full paper to learn more!
Updates to placeholder symbols
The HGNC continues to update placeholder symbols whenever new data becomes available. In the past few months we have updated the following genes based on discussions between an HGNC curator and researchers working on the gene:
- C8orf37 -> CFAP418, cilia and flagella associated protein 418
- FAM155A -> NALF1, NALCN channel auxiliary factor 1
- FAM155B -> NALF2, NALCN channel auxiliary factor 2
The following genes have been renamed following updates to their annotation models, resulting in a change in locus type from protein coding to long non-coding RNA:
- C17orf77 -> CD300LD-AS1, CD300LD antisense RNA 1
- C9orf147 -> HSDL2-AS1, HSDL2 antisense RNA 1
- C9orf106 -> LINC02913, long intergenic non-protein coding RNA 2913
- C14orf177 -> LINC02914, long intergenic non-protein coding RNA 2914
- C15orf54 -> LINC02915, long intergenic non-protein coding RNA 2915
- C11orf72 -> NDUFV1-DT, NDUFV1 divergent transcript
New gene groups
Here are some examples of new gene groups that we have made within the last couple of months:
- Methylcrotonyl-CoA carboxylase subunits (MCCC)
- Transcription factor AP-2 family (TFAP2)
- Tet methylcytosine dioxygenase family (TET)
- Adducin family (ADD)
- TNRC6 adaptor family (TNRC6)
- PARN exonuclease family
- TLDc domain containing (TLDC)
Gene Symbols in the News
Babies born with spinal muscular atrophy in the UK can now benefit from gene therapy with an active copy of the gene SMN1. This therapy has been heralded as the ‘most expensive’ drug treatment ever approved and has already been available in other countries, such as the USA. It requires just a single treatment early on in life.
In cancer-related news, the cell surface-expressed gene LRRN4CL has been identified as a biomarker for melanoma following a CRISPR activation screen. There is hope that a drug could be developed in the future to target the LRRN4CL protein in melanoma patients, particularly as it is a cell surface protein and therefore drugs could be extracellular.
Most humans carry a pseudogenised copy of the SIGLEC12 gene, but about 30% of people have a protein coding version. The protein version has been suggested to partially explain why humans have high rates of carcinoma compared to other great apes, as the protein appears to be involved in aberrant cell signalling and its expression has been associated with poor prognosis in colorectal cancer patients.
In COVID-19 news, variants in the following genes have been newly associated with an increased risk of contracting the disease: ERMP1, FCER1G and CA11. The same study corroborated previously-reported variants in the ABO and SLC6A20 genes. Additionally, the study identified variants in the IL10RB, IFNAR2 and OAS1 genes that are linked to patients suffering from a more severe form of the disease.
Finally, we bring you news about a dog gene! A study from Finland has identified the causal gene for nonsyndromic early-onset hereditary hearing loss in Rottweilers as LOXHD1. We have already approved this gene symbol for the dog gene in VGNC via the pipeline described in the HCOP section above. The human ortholog has also been associated with deafness.
Meeting News
Elspeth attended the virtual NC-IUPHAR meeting on the 15th and 16th April. While it was enjoyable, we’re looking forward to visiting Paris for a future meeting!
Publications
Yates B, Gray KA, Jones TEM, Bruford EA. Updates to HCOP: the HGNC comparison of orthology predictions tool. Brief Bioinform. 2021 May 6. PMID: 33959747 DOI: 10.1093/bib/bbab155
