Winter newsletter 2022
Newsletters ·
Thanks to our Scientific Advisory Board
We would like to thank all the members of our SAB for attending our (mostly virtual) annual meeting from 27-28th January. We hope that we might be able to host a less virtual version in the future! We were pleased to welcome our new board member, Cecilia Arighi, who works within several different protein information projects, including UniProt, Protein Ontology and BioCreative. While Professor Helen Firth has rotated off the board, we are delighted that she has agreed to remain associated with HGNC as our Clinical Advisor.
Play ‘Genele’ - the gene symbol guessing game
How many of you have tried Genele, the fun gene symbol guessing game made by Dr Andrew Holding? It might just remind you of a popular word guessing game that has taken the world by storm in the last few months ;-). We love Genele so much that we have linked to it from our home page. So take a break for a few minutes daily to guess the gene symbol - go on, you can even use genenames.org to help you out a little!
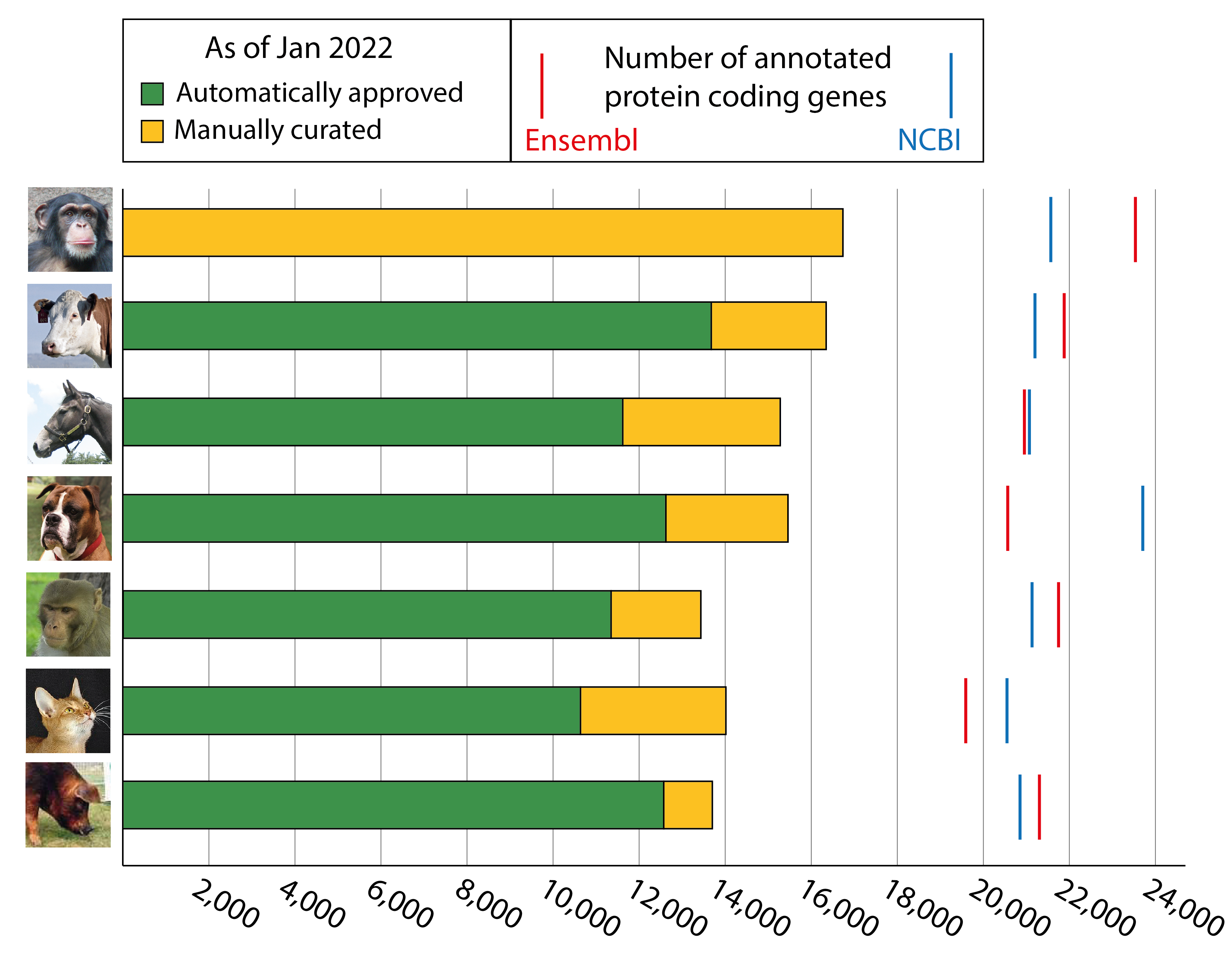
Progress on the VGNC project
We now have over ~108,500 genes approved in total for our VGNC project, with 14,019 cat genes, 16,751 chimpanzee genes, 16,856 cattle genes, 15,567 dog genes, 15,704 horse genes, 14,677 Rhesus macaque genes and 13,898 pig genes with approved VGNC symbols. This includes over 6,000 new VGNC genes within the last year! Below is a graph showing our progress with each core species.
Wanted - a new VGNC full stack developer
We are currently advertising for a new full stack developer to work on our VGNC project. The closing date is 10th March, so please notify anybody you know that might be interested!
Update on genes with the ‘stable’ tag
We have 2821 gene symbols tagged as ‘stable’ as of February 23rd 2022, an increase of 209 since our Autumn newsletter. Examples of genes within the new stable set include NHEJ1, the causative gene for Severe combined immunodeficiency with microcephaly growth retardation, and sensitivity to ionizing radiation, NKX6-2, the causative gene for Spastic ataxia 8, autosomal recessive, with hypomyelinating leukodystrophy and KNG1, the causative gene for High molecular weight kininogen deficiency.
In order for a gene to be marked as stable, an HGNC curator manually reviews the gene symbol to check that this is unlikely to ever be changed in future. During the review of the 209 genes that were marked as stable in the last quarter, no gene symbols were changed and the descriptive names of just seven genes were altered. In most cases this was to make the name more functionally informative - the gene name of NUBPL was updated from “nucleotide binding protein like” to “NUBP iron-sulfur cluster assembly factor, mitochondrial”; EPG5 was updated from “ectopic P-granules autophagy protein 5 homolog” to “ectopic P-granules 5 autophagy tethering factor”; CD3E was updated from “CD3e molecule” to “CD3 epsilon subunit of T-cell receptor” and CD3G was updated from “CD3g molecule” to “CD3 gamma subunit of T-cell receptor complex”.
Updates to placeholder symbols
Since the last newsletter we have continued to work on updating C#orf symbols where possible. The following symbols were all updated based on data in publications:
- C11orf49 -> CSTPP1, centriolar satellite-associated tubulin polyglutamylase complex regulator 1
- C7orf26 -> INTS15, integrator complex subunit 15
- C9orf116 -> PIERCE1, piercer of microtubule wall 1
- C15orf65 -> PIERCE2, piercer of microtubule wall 2
- C7orf61 -> SPACDR, sperm acrosome developmental regulator
The nomenclature of the FAM71 family was updated following a consultation with researchers working on these genes. The symbol “GARI” had been published for FAM71F2 but this was not a suitable gene symbol for approval because it is a poor search term and is similar to the approved symbol GAR1. The community agreed upon “GARIN” for “golgi associated RAB2 interactor” as a suitable alternative root symbol for the FAM71 family. For genes where the function of the encoded protein has not been confirmed as a golgi associated RAB2 interactor, the term “family member” was added into the gene name. The updates were as follows:
- FAM71F2 -> GARIN1A, golgi associated RAB2 interactor 1A
- FAM71F1 -> GARIN1B, golgi associated RAB2 interactor 1B
- FAM71D -> GARIN2, golgi associated RAB2 interactor 2
- FAM71B -> GARIN3, golgi associated RAB2 interactor 3
- FAM71A -> GARIN4, golgi associated RAB2 interactor family member 4
- FAM71E1 -> GARIN5A, golgi associated RAB2 interactor 5A
- FAM71E2 -> GARIN5B, golgi associated RAB2 interactor family member 5B
- FAM71C -> GARIN6, golgi associated RAB2 interactor family member 6
Gene Symbols in the News
We bring news of two gene therapy stories, the first is the successful treatment of sickle cell disease via gene editing of the BCL11A gene to switch adults to making fetal hemoglobin rather than the affected adult hemoglobin that causes the red blood cells to ‘sickle’. The treatment is so successful that patients have not needed hospital visits since. The second is in earlier stages and is the first attempt to treat Tay-Sachs disease, caused by mutation of the HEXA gene. A low dose of treatment was delivered to the brain and spine of two young girls, who are now clinically stable with slowed or no disease progression. Further studies will use higher doses of treatment.
A new as yet unnamed neurodevelopmental disorder has been identified that is caused by mutation of the PAX5 gene. After discovering a mutation in one patient, researchers used GeneMatcher to find another 15 patients with PAX5 mutations who all exhibited similar characteristics of developmental delay, intellectual disability and autism spectrum disorder.
An SNP located within a chemokine receptor cluster that affects expression of CCR1, CCR2, CCR3 and CCR5 has been shown to increase the risk of severe COVID-19 disease but lower the risk of becoming infected with HIV. Interestingly, this SNP was inherited from Neanderthals and rose in frequency over 10,000 years ago, suggesting that this may have been due to selective pressure from a virus infecting humans around this time, such as smallpox.
Finally we bring news of a gene variant identified in dog that is responsible for the small size in breeds such as chihuahua and miniature schnauzers. The mutation is carried on a long non-coding RNA gene that is antisense to the protein coding IGF1 gene.
Publications
Braschi B, Omran H, Witman G.B, Pazour, G.J, Pfister, K.K, Bruford, E.A, King, S.M. Consensus nomenclature for dyneins and associated assembly factors. J Cell Biol. 2022 Feb 7;221(2):e202109014. Epub 2022 Jan 10. PMID: 35006274 PMCID: PMC8754002 DOI: 10.1083/jcb.202109014. Please also read the accompanying guest blog post by Dr Stephen M. King Naming Dynein Components and their Cytoplasmic Associated Factors.
Ho M, Thompson B, Fisk JN, Nebert DW, Bruford EA, Vasiliou V, Bunick CG. Update of the keratin gene family: evolution, tissue-specific expression patterns, and relevance to clinical disorders. Hum Genomics. 2022 Jan 6;16(1):1. DOI: 10.1186/s40246-021-00374-9. PMID: 34991727. PMCID: PMC8733776
