Autumn newsletter 2021
Newsletters ·
New links to GenCC
We are pleased to announce that HGNC now displays links from our Symbol Reports to curated gene-disease relationships in the GenCC (The Gene Curation Coalition) database. The HGNC is a member of the GenCC project that brings together multiple groups that are either directly involved in gene-disease curation or that promote standards needed to support this curation. Genes reported as being associated with disease by the GenCC project are on the HGNC’s priority list for gene symbol stabilisation.
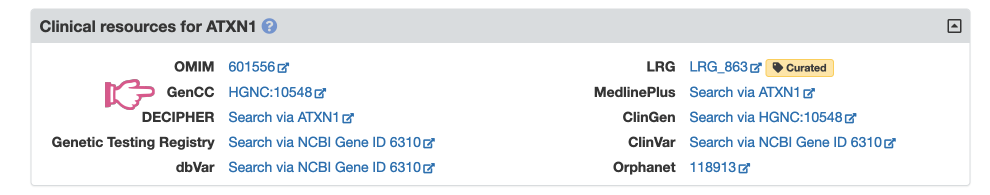
The new links to GenCC can be found in the Clinical Resources section of our Symbol Reports, as shown for the ATXN1 Symbol Report below:
Recommendations for reporting gene fusions
HGNC has worked with the scientific community to determine a new standard for describing human fusion genes - recommending the use of a double colon (::) to separate the two gene symbols involved in the gene fusion, e.g., BCR::ABL1. You can read our new article, “HUGO Gene Nomenclature Committee (HGNC) recommendations for the designation of gene fusions” for full details as to why the double colon was chosen. In brief, gene fusions in the majority of publications have been denoted by use of either a hyphen (-) or a forward slash (/), but these two characters can result in confusion as they are used to denote many other concepts, such as readthrough transcripts and pathway descriptions.
As usual, the HGNC has been collaborating and communicating with as many other groups and resources as possible to ensure that this standardisation is supported and applied across the biomedical sphere. The new recommendation for use of the double colon is supported by:
- HGVS (Human Genome Variation Society)
- ISCN (International System for human Cytogenetic Nomenclature)
- WHO Classification of Tumors
- COSMIC
- OMIM
- Atlas of Genetics and Cytogenetics in Oncology and Haematology
- Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer
- Tumor Fusion Gene Data Portal
Update on genes with the ‘stable’ tag
HGNC curators have been working hard to continue adding the ‘stable’ tag to genes in the last quarter. As of November 19th 2021 we have 2612 stable symbols - an increase of 169 since our previous newsletter. As described in our September blog post Stability in the time of COVID-19, we have recently been prioritising review of the nomenclature of genes linked to COVID-19. Unsurprisingly the number of publications associated with these genes has greatly increased since the beginning of 2020; for example, the symbol ACE2 appears in PubMed 3586 times in 2020 and 3941 times so far in 2021, compared to 157 times in 2018 and 173 times in 2019. We used the COVID-19 UniProtKB webpage as our source of human genes related to COVID-19. The 81 proteins encoded by the human genome listed on that webpage include immune related genes, transcription factors, enzymes and receptors: we have been able to add the stable tag to over 85% of the relevant gene symbols. The symbol stability review has resulted in the change of just one gene symbol in the last quarter - since the publication of the blog post we have now updated PHB (HGNC:8912) to the more search-friendly symbol PHB1, with the added advantage that the human gene symbol is now consistent with the yeast PHB1 and nematode worm phb-1 gene symbols.
Updates to placeholder symbols
Over the last three months the HGNC has been able to update a number of C#orf symbols to symbols with the root CFAP# (based on the identification of the gene as the ortholog of a gene with the FAP#, flagellar associated protein, nomenclature in Chlamydomonas), there has been a further update based on the identification of the gene product as part of a complex, and one based on a change of locus type from protein coding to long non-coding RNA:
- C9orf135 -> CFAP95, cilia and flagella associated protein 95
- C1orf158 -> CFAP107, cilia and flagella associated protein 107
- C1orf189 -> CFAP141, cilia and flagella associated protein 141
- C1orf194 -> CFAP276, cilia and flagella associated protein 276
- C5orf51 -> RIMOC1, RAB7A interacting MON1-CCZ1 complex subunit 1
- C11orf45 -> KCNJ5-AS1, KCNJ5 antisense RNA 1
New gene groups
We have worked on our Nuclear hormone receptors gene group hierarchy to make the subgroups consistent with the IUPHAR/BPS Guide to Pharmacology nuclear hormone receptor pages. This group now has the following subgroups (as displayed on the Nuclear hormone receptors gene group page):

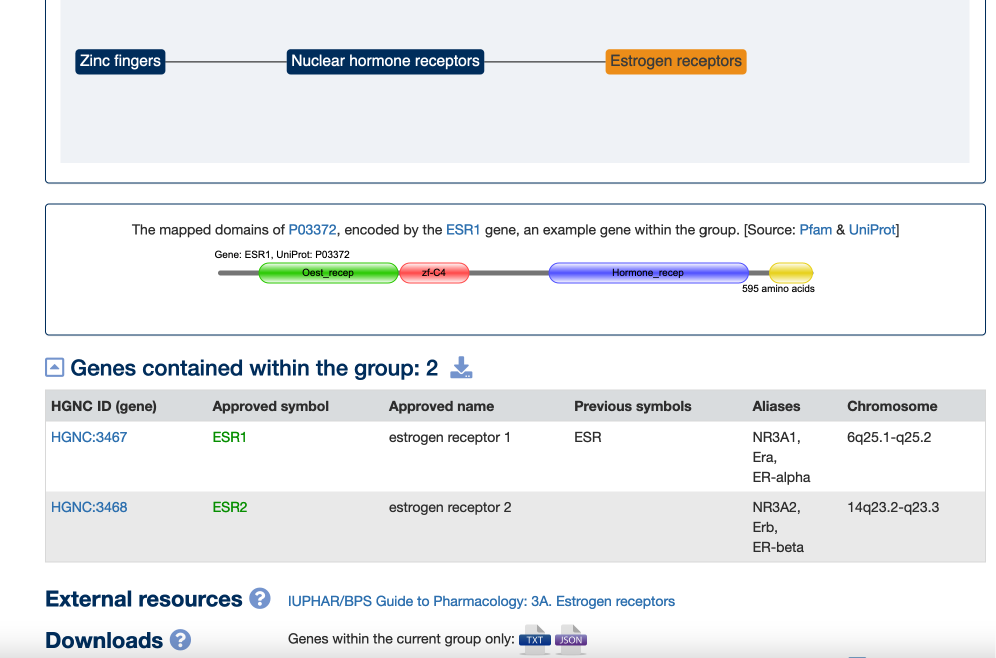
An example subgroup, Estrogen receptors (ESR), is shown below. The link to the appropriate IUPHAR page is found towards the bottom of the Gene group report:
Gene Symbols in the News
A new study may explain why certain apes, including humans, do not have tails - these apes have an Alu element insertion within a noncoding region of the TBXT gene, while this insertion is not found in primates that have tails.
A study has identified the olfactory receptor that is necessary for us to detect caramel type smells. The OR5M3-encoded receptor is activated by only two odorants, furaneol, which allows us to smell caramel notes in food such as coffee and strawberries, and homofuraneol, which gives a caramel-type smell to durian and other fruits.
In many parts of the world humans are growing taller and reaching puberty at an earlier age. New research pins the MC3R gene, which encodes a receptor expressed in the brain, as the link between accessibility of food and this accelerated growth and development.
In COVID-19 news, a variant of the previously little-studied LZTFL1 gene, which is present in a higher proportion in people of South Asian descent, has been associated with a doubling in the risk of death from COVID-19. The current hypothesis for this association is that the risky LZTFL1 variant prevents a protective mechanism in lung cells that would reduce the amount of ACE2 protein on the lung cell surface.
A different gene, APOL1, has been associated with more severe COVID-19 disease in African Americans, based on a study of patients admitted to the Hospital of the University of Pennsylvania. The same gene variant was also linked to increased risk of developing sepsis.
A separate study on young, critically ill patients with COVID-19 who had no known underlying health conditions used machine learning to identify genes that were more active in these patients. One of the identified genes was ADAM9; in vitro studies suggest that higher levels of the ADAM9 gene product is associated with an increase in SARS-CoV-2 virus duplication.
Meeting News
Elspeth attended the virtual Autumn NC-IUPHAR meeting on the 19th November. On the same day, Ruth attended the annual virtual RNAcentral consortium meeting.
Publications
Braschi B, Seal RL, Tweedie S, Jones TEM, Bruford EA. The risks of using unapproved gene symbols. Am J Hum Genet. 2021 Oct 7;108(10):1813-1816. DOI:10.1016/j.ajhg.2021.09.004 PMID:34626580 Please also see our companion blog post to this article “The risks of using unapproved symbols”.
Bruford EA, Antonescu CR, Carroll AJ, Chinnaiyan A, Cree IA, Cross NPC, Dalgleish D, Gale RP, Harrison CJ, Hastings RJ, Huret JL, Johansson B, Le Beau M, Mecucci C, Mertens F, Verhaak R, Mitelman F. HUGO Gene Nomenclature Committee (HGNC) recommendations for the designation of gene fusions. Leukemia. 2021 DOI: 10.1038/s41375-021-01436-6 PMID:34615987 There is an accompanying Editorial for this article: Gale RP, Hochhaus A, Cross NCP, Harrison CJ. HGNC nomenclature for fusion genes. Leukemia. 2021 DOI: 10.1038/s41375-021-01437-5 PMID: 34611300
